Figure 2.
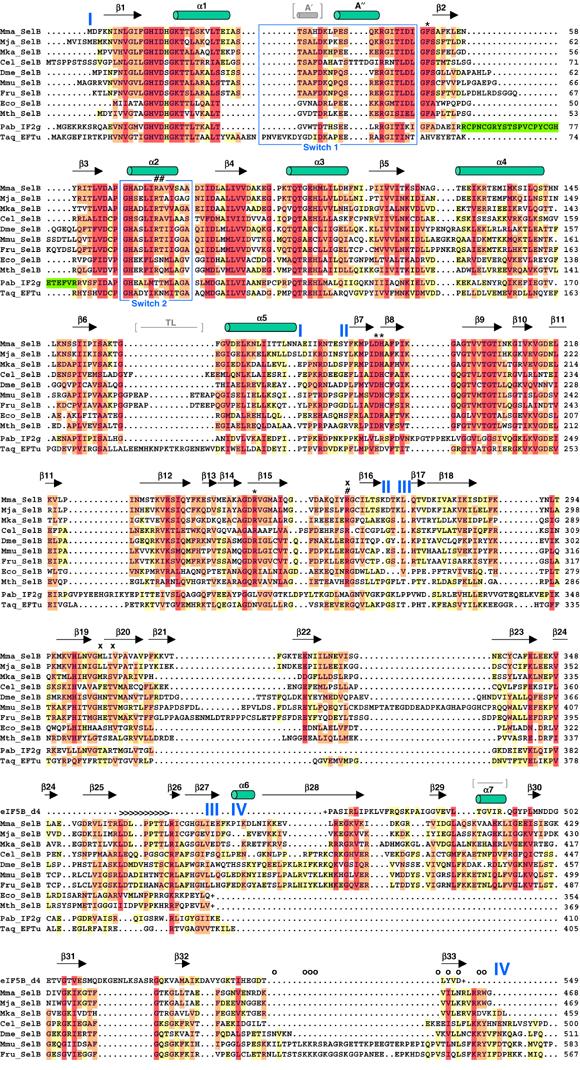
Secondary structure of SelB:GDP and structure-based alignment of selected SelB sequences from all kingdoms. Different SelB sequences from archaea, bacteria and eukaryotes were obtained from the NCBI protein sequence database (www.ncbi.nlm.nih.gov/Entrez/) and are depicted as follows: Mma, M. maripaludis; Mja, M. jannaschii; Mka, Methanopyrus kandleri; Cel, Caenorhabditis elegans; Dme, Drosophila melanogaster; Mmu, Mus musculus; Fru, Fugu rubripes; Eco, E. coli and Mth, M. thermoacetica. Domain borders are indicated in blue. Bacterial SelBs are truncated (depicted with (+)) after domain III due to the completely unrelated fold of domain IV. The alignment was performed using CLUSTAL X (Thompson et al, 1997) and edited manually with GeneDoc (www.psc.edu/biomed/genedoc). The colouring is according to the Gonnet PAM 250 series with 40% (yellow), 70% (orange) and 100% similarity (red). For comparison, the sequences of Thermus aquaticus EF-Tu (Taq), Pyrococcus abyssi IF2γ (Pab) and the core part of Methanobacterium thermoautotrophicum eIF5B domain IV are also aligned. The numbering of these sequences is according to the published structures. The zinc knuckle insertion in IF2γ domain I is depicted in green. Several important SelB residues are labelled according to their possible function: 5′ phosphate recognition (#), blocking of the 5′ recognition (x), aminoacyl binding (*) and tRNA backbone contacts (>). Mutations in residues affecting the interaction of murine SelB with SBP2 are depicted with (o). The Switch 1 and 2 regions are boxed in blue. The additional A′ helix and the thermophile loop (TL) in EF-Tu and the loop in eIF5B domain IV replaced by α7 in SelB are indicated in grey, between brackets.
