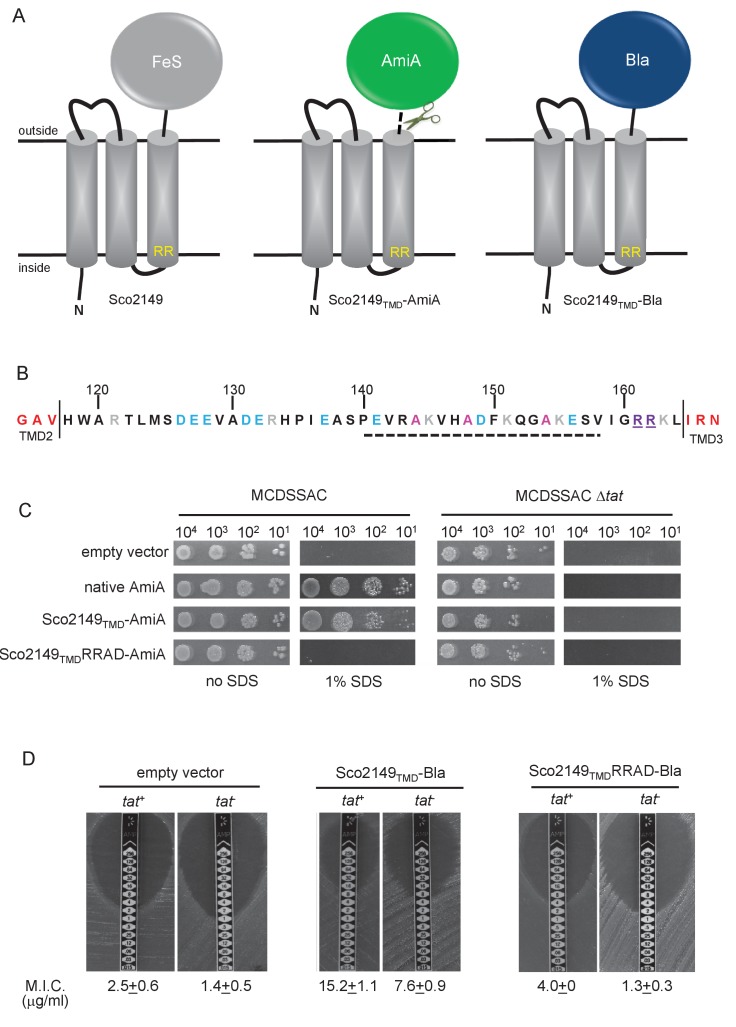
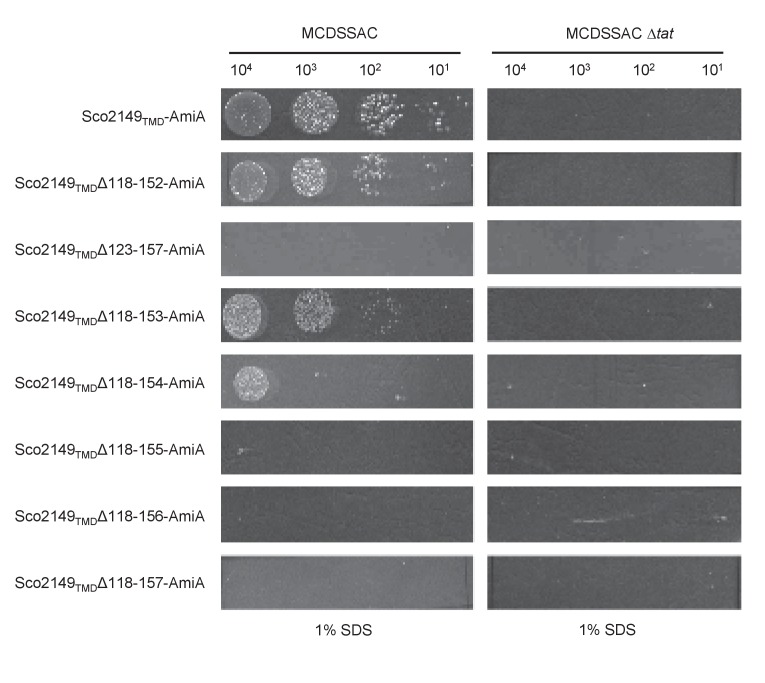
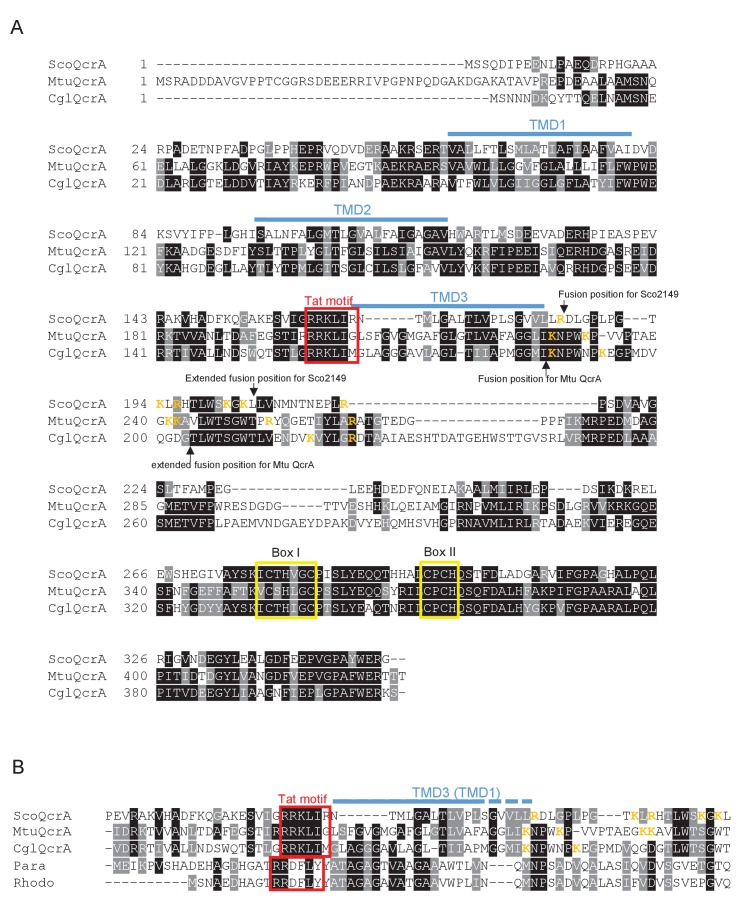
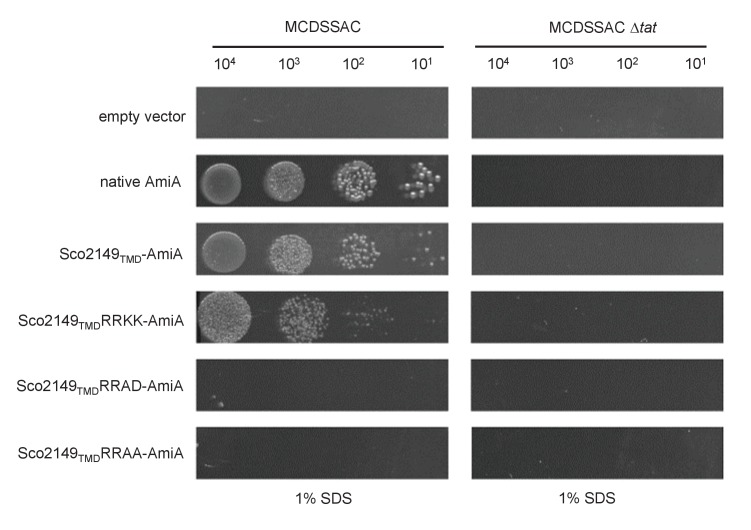
Figure 1. Sco2149TMD-reporter fusions to follow membrane insertion.
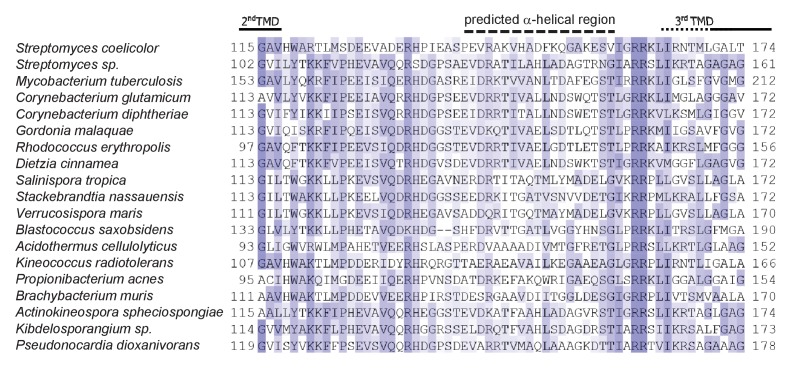
(A) Cartoon representations of the S. coelicolor Rieske protein, Sco2149, and the Sco2149TMD-AmiA and Sco2149TMD-Bla fusions. A signal peptidase I cleavage site (indicated by scissors) was introduced between the end of TMD3 and the AmiA sequence to allow release of AmiA from the membrane (Keller et al., 2012). The position of the twin-arginine motif is indicated by RR. (B) Sequence of the Sco2149 cytoplasmic loop region between TMDs 2 and 3. Amino acids predicted to be part of TMDs 2 and 3 are shown in red. The twin arginines of the Tat recognition motif are given in purple underline. Predicted α-helical secondary structure is shown with a dotted line, and alanine residues within this region that were mutated to proline are shown in pink. Negatively charged amino acids in the loop region are shown in blue, positively charged ones in grey. (C) E. coli strain MCDSSAC (which carries chromosomal deletions in the signal peptide coding regions of amiA and amiC) or an isogenic tatABC mutant containing either pSU-PROM (empty vector), or pSU-PROM producing native AmiA, Sco2149TMD-AmiA or a variant where the twin-arginines were substituted to AD (Sco2149TMDRRAD-AmiA), were spotted, after serial dilution, on LB medium in the absence or presence of 1% SDS. The plates were incubated for 20 hr at 37°C. (D) Representative images of M.I.C.Evaluator strip tests of strains MC4100 (tat+) and DADE (tat-) harbouring pSU-PROM (empty vector), pSU-PROM Sco2149TMD-Bla or pSU-PROM Sco2149TMDRRAD-Bla are shown. The mean M.I.C ± s.d. for strains harbouring these constructs is given at the bottom of each test strip (where n = 4 biological replicates for each strain harbouring the empty vector, n = 5 biological replicates for each strain harbouring pSU-PROM Sco2149TMD-Bla and n = 3 biological replicates for each strain harbouring pSU-PROM Sco2149TMDRRAD-Bla).