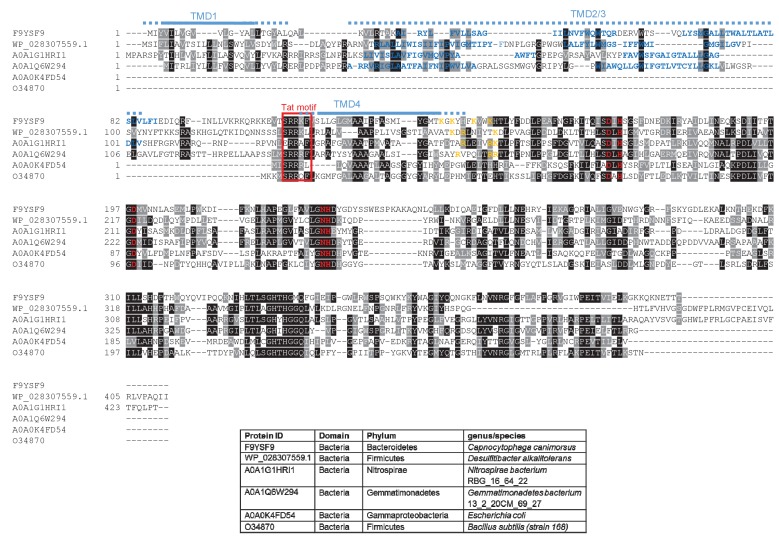
Figure 7. Sequence alignment of selected YkuE-related proteins.
Sequences of polytopic YkuE-like metallophosphoesterase proteins from the indicated prokaryotes were aligned, alongside the shorter homologues from E. coli and B. subtilis using ClustalW (http://www.ch.embnet.org/software/ClustalW.html) and Boxshade (http://www.ch.embnet.org/software/BOX_form.html). Predicted positions of the TMDs (using the SCAMPI2/TOPCONS servers (Tsirigos et al., 2015, 2016)) are shown in blue. Note that residues in predicted TMD2 and TMD3 are not well aligned across the homologues and therefore amino acids predicted to be in TMD2 and TMD3 for each protein are individually marked in blue font. Positively charged amino acids immediately downstream of TMD5 are shown in orange. The consensus twin arginine motif is boxed in red and amino acids predicted to coordinate the metal ion cofactor are shown in red font.