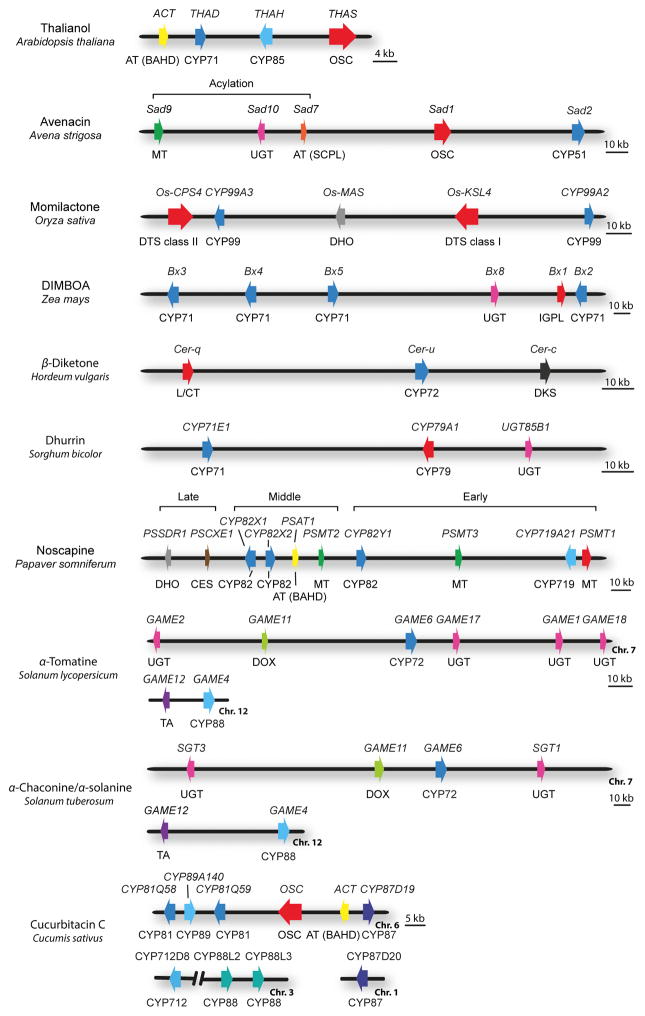
Fig. 2.
Genomic organization of plant metabolic gene clusters. Examples that exemplify different cluster types and biosynthetic pathways are shown. The genes are indicated by arrows. The gene(s) for the first committed pathway step are indicated in red. Gene names are indicated above the clusters, and class of biosynthetic enzyme below. Abbreviations: OSC, oxidosqualene cyclase; DTS, diterpene synthase (class I and class II shown); IGPL, indole 3-glycerol phosphate lyase; DKS, diketone synthase; AT (BAHD), BAHD-acyltransferase; AT (SCPL), SCPL-acyltransferase; MT, methyltransferase; UGT, UDP-dependent sugar transferase, DHO, dehydrogenase/reductase; L/CT, lipase/carboxyltransferase; CES, carboxylesterase; DOX, dioxygenase; TA, transaminase; CYP, cytochrome P450. The maize DIMBOA pathway includes three genes that are not shown in the figure: Bx7, which is separated from the core cluster by an intervening region of 15 Mb; the sugar transferase gene Bx9, which is located on a different chromosome; and finally Bx6, which is not shown because its genomic location has not yet been established. [Correction added after online publication 26 April 2016; β-Diketone Hordeum vulgaris cluster has been corrected.]