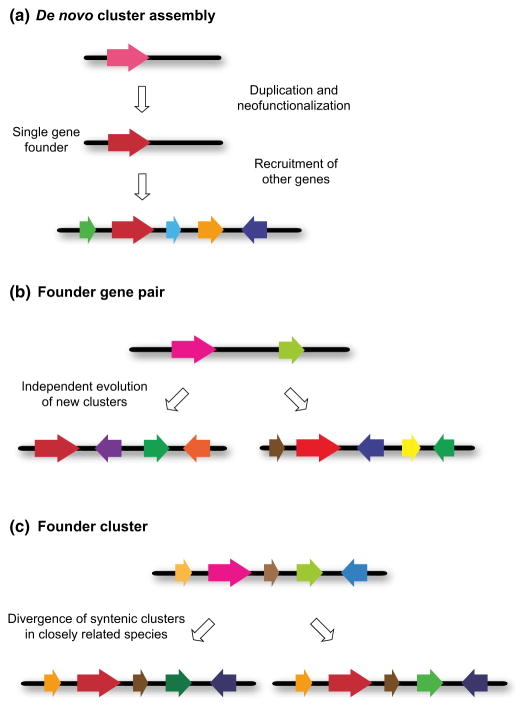
Fig. 6.
Origins of plant metabolic clusters. The scenarios shown are based on current knowledge of characterized plant metabolic clusters (see text). (a) De novo cluster assembly. The first step in this process is the recruitment of the gene for the first committed step in the pathway by gene duplication and neofunctionalization. This gene then ‘seeds’ the formation of a multigene cluster for a new metabolic pathway by an as yet unknown mechanism. (b) Formation of new metabolic clusters from a founder gene pair template. In this scenario, an ancestral gene pair (e.g. encoding a terpene synthase and a cytochrome P450 (CYP) enzyme) may give rise to different metabolic clusters for new natural product pathways as a result of independent events involving gene rearrangement and recruitment of new genes. This is exemplified by the Arabidopsis thaliana thalianol and marneral clusters (Field et al., 2011). These are both triterpene pathways, but the scaffolds are not the same and the subsequent modifications are different. For both (a) and (b) the process of cluster formation will be accompanied by refinement and optimization of individual pathway components and establishment of a functional gene neighbourhood that allows for coordinate pathway control. (c) Diversification of an ancestral founder cluster in syntenic regions of closely related taxonomic lineages. In these cases, these pathways may have a common scaffold but make slightly different compounds.