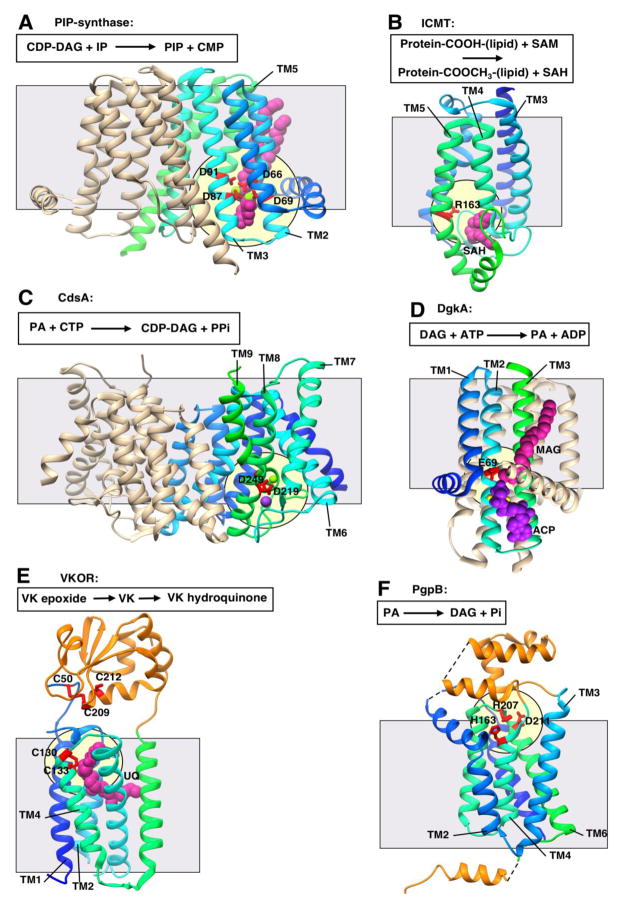
Figure 2. Representative structures showing Interfacial catalysis (I).
All structures are shown in ribbon representation with bound ligands shown in CPK representation. Transmembrane domains are shown in gradient coloring from the N-terminus to the C-terminus (blue to cyan to green) while extramembrane soluble domains are shown in orange. Grey square shows approximate location of the membrane border. Pale yellow circle indicates location of the putative active site. All structures are oriented with the cytosolic face of the membrane on the bottom. A schematic of the reaction catalyzed is shown above each structure. A. Phosphatidylinositol-phosphate synthase (PIP-synthase; PDB 5D92) with four aspartate residues from the CDP-AP signature sequence in red responsible for metal coordination and catalysis. Magnesium ion in light green and lipid CDP-diacylglycerol (CDP-DAG) in magenta. CDP-DAG, cytidine diphosphate diacylglycerol; IP, inositol-phosophate; PIP, phosphatidylinositol-phosphate; CMP, cytidine monophosphate. B. Isoprenylcysteine carboxyl methyltransferase (ICMT; PDB 4A2N) with putative catalytic residue in red. Methyl donor product S-Adenosyl-homocysteine (SAH) in magenta. SAM, S-Adenosyl methionine C. CDP-DAG synthetase (CdsA; PDB 4Q2E) with putative metal binding and catalytic residues in red. Magnesium ion in light green. Potassium ion in purple. PA, phosphatidic acid; CTP, cytidine triphosphate. D. Diacylglycerol kinase (DgkA; PDB 4UXX) with putative catalytic residue in red. Monoacylglycerol (MAG) in magenta, adenylmethylenediphosphonate (ACP) in purple, two zinc ion in yellow (partially obscured by ACP). E. Vitamin K epoxide reductase (VKOR; PDB 3KP9) cysteines proposed in electron transfer in red. Ubiquinone (UQ) in magenta. VK, vitamin K. F. Phosphatidylglycerolphosphate phosphatase B (PgpB; PDB 4PX7) with putative catalytic residues in red.