Fig. 1.
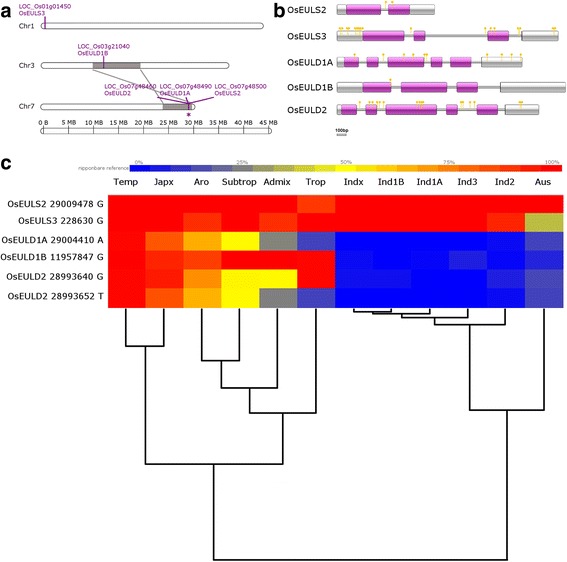
Comparison of DNA sequences encoding EUL homologs from O. sativa. a. Distribution and duplication of EUL genes in the Oryza sativa subsp. japonica genome. Sequences for the different EULs were mapped on the chromosomes based on their start position and annotated with their locus number. Blocks containing EUL genes resulting from segmental duplication events are indicated in gray and connected by lines. Tandem duplications are indicated by an asterisk. The chromosome map was prepared using MapChart and drawn to scale. b. Intron-exon structure of the EUL genes. Shown in white are the 5′ and 3′ UTRs, exons are shown in purple. Introns are represented by grey bars between the exons. SNPs identified by SNP-seek are indicated in yellow.c. Distribution of SNP alleles in different rice varieties. The percentage of the Nipponbare reference allele in the different subvarieties is indicated by the heat map. The dendrogram is generated by DendroUPGMA (http://genomes.urv.cat/UPGMA/) (Garcia-Vallve et al. 1999)
