Figure 2.
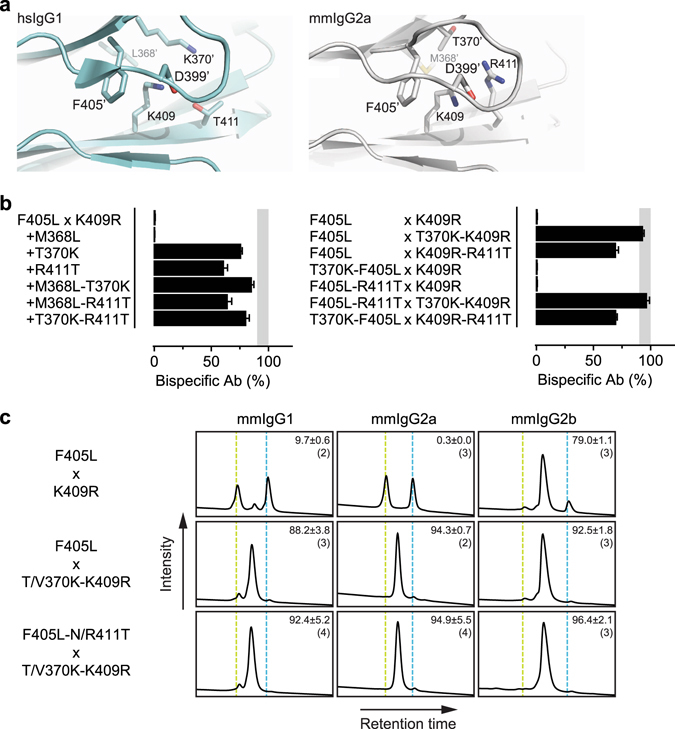
(a) Amino acid residues surrounding K409 at the CH3-CH3 interface of hsIgG1 (left panel) and mmIgG2a (right panel). Images were generated from PDB IDs 3AVE54 (hsIgG1) and 1IGT61 (mmIgG2a) using PyMOL software (Schrödinger). ‘Indicates residues on opposite CH3 domain. (b) Efficiency of cFAE as measured by HIC of symmetric (left panel) and asymmetric (right panel) mixtures of 2F8- and 7D8-derived chimeric parental antibodies. The chimeric parental antibodies contained human variable regions and human IgG1 constant regions, wherein only the CH3 domain was swapped with that of mouse IgG2a and containing the indicated point mutations. Data represent mean ± SEM of at least two separate experiments. Shaded area represents 90–100% efficiency. (c) Exemplary Hydrophobic Interaction Chromatography (HIC) profiles of bispecific antibodies generated by cFAE using combinations of 2F8- and 7D8-derived chimeric parental antibodies. The chimeric parental antibodies contained human variable regions and mouse constant regions that contained the indicated point mutations (above and left of panels, respectively). Numbers represent mean ± SEM (n) percentage of bispecific antibody product (middle peaks). Vertical lines correspond with the retention times of the individual 2F8 (green) and 7D8-derived (blue) parental antibodies.
