Figure 7.
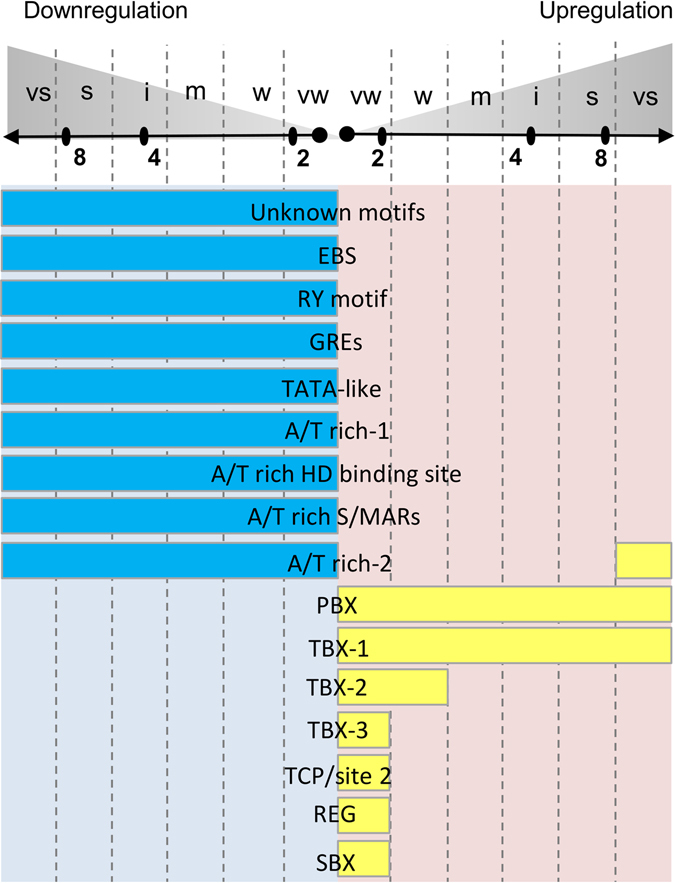
cis-Regulatory motifs found enriched in the upstream regions of the genes responded to auxin within certain fold change intervals. Unknown motifs (atacta/tagtat, atagta/tactat; atatga/tcatat, atatgt/acatat, taatag/ctatta, tatgta/tacata); EIN3-binding site EBS (atacat/atgtat)30; B3-family binding RY motif (tgcatg/catgca)31; bZIP-binding GRE/ABRE motifs (tacgtg/cacgta; acacgt/acgtgt, acgtgg/ccacgt)32, TATA-like (aatata/tatatt, actata/tatagt, atataa/ttatat, atatac/gtatat, atatag/ctatat, tataac/gttata, tatata, tatatg/catata; atatat)33; A/T rich-1 (aataat/attatt, aattat/ataatt, ataata/tattat, attaat, attata/tataat, taaata/tattta, tattaa/ttaata); A/T-rich Homeodomain (HD) binding site (taatta)34; A/T-rich S/MARs (aatatt)35; A/T rich-2 (atatta/taatat); PBX (atgggc/gcccat); TBX-1 (ttaggg/ccctaa), TBX-2 (aaccct/agggtt, acccta/tagggt), TBX-3 (aaaccc/gggttt, ctaggg/ccctag, SBX (aagccc/gggctt)17; TCP/site216, 21 (agccca/tgggct, tgggcc/ggccca); REG (aggccc/gggcct)17, 18. For more detailed descriptions see Supplementary Table 8.
