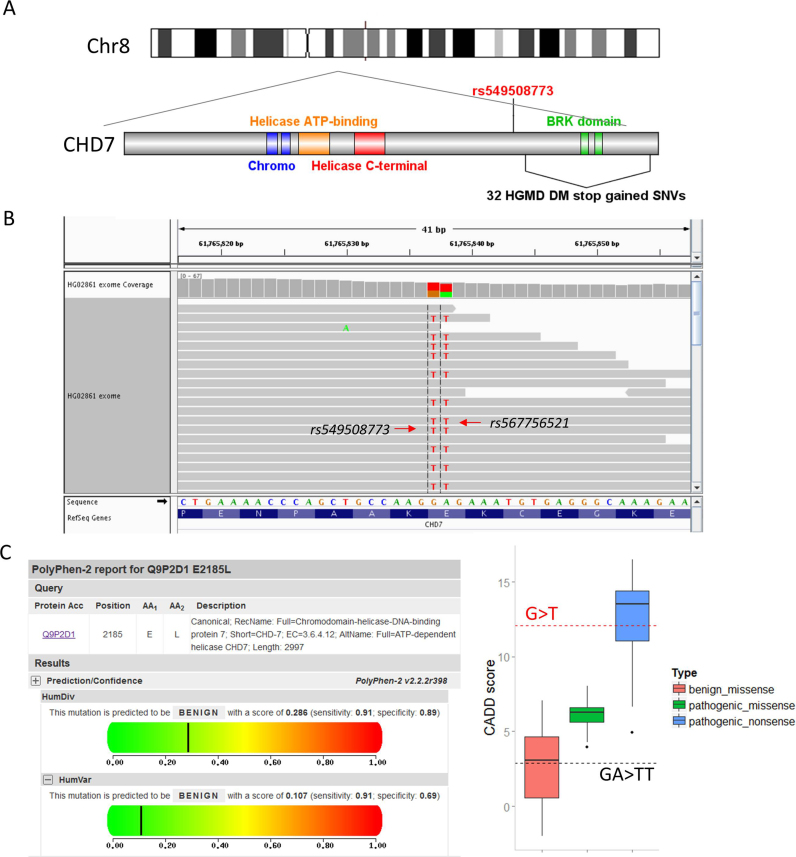
Figure 3.
A pathogenic SNV is rescued in its specific genomic context. (A) Compared with SNV rs549508773 (red), 32 disease-causing (DM) stop-gained SNVs (black) recorded in the HGMD database are located downstream of the gene CHD7. (B) The figure shows an IGV image of the whole exome sequencing data of HG02861, a healthy participant in the 1000 Genomes Project. As shown in the figure, SNV rs567756521 rescues the stop-gained mutation, and the combined effect is a single amino acid substitution (Glu>Leu). (C) Polyphen-2 predicted the single amino acid (Glu>Leu) as a benign mutation (Left). Boxplot of CADD scores for three different kinds of mutations (benign missense, pathogenic missense and pathogenic nonsense) collected from the CHD7 database (Right). The score for rs549508773 (G>T) is located within the range of pathogenic nonsense (red dotted line). The score for the MNV (GA>TT) is located within the range of benign missense (black dotted line).