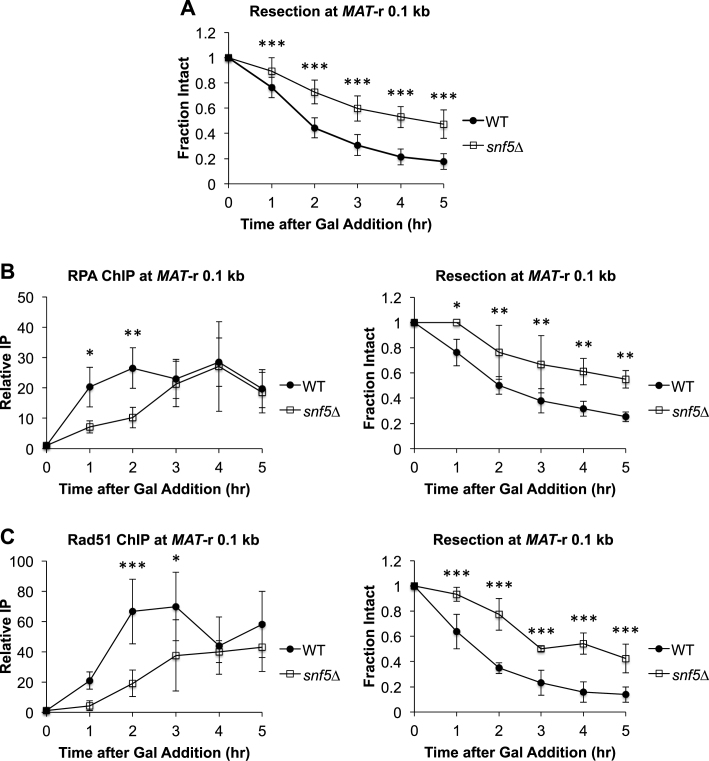
Figure 1.
Initiation of DNA end resection at MAT is impaired in snf5Δ cells. OK (A) Asynchronous WT (n = 9) and snf5Δ (n = 10) cells were harvested at 1 h intervals after addition of galactose to induce a MAT DSB. Resection was monitored by qPCR with primers that anneal 0.1 kb to the right of the MAT DSB. (B) Recruitment of RPA 0.1 kb to the right of the MAT DSB was monitored by ChIP (left) and DNA end resection was simultaneously monitored (right) in WT (n = 3) and snf5Δ (n = 3) asynchronous cells after addition of galactose to induce a MAT DSB. (C) Recruitment of Rad51 0.1 kb to the right of the MAT DSB was monitored by ChIP (left) and DNA end resection was simultaneously monitored (right) in WT (n = 3) and snf5Δ (n = 3) asynchronous cell populations as described in B. Error bars denote one standard deviation. Statistical differences between WT and snf5Δ at time points were assessed by two-way ANOVA with Holm–Sidak post-hoc analysis. *P < 0.05, **P < 0.01, ***P < 0.001.