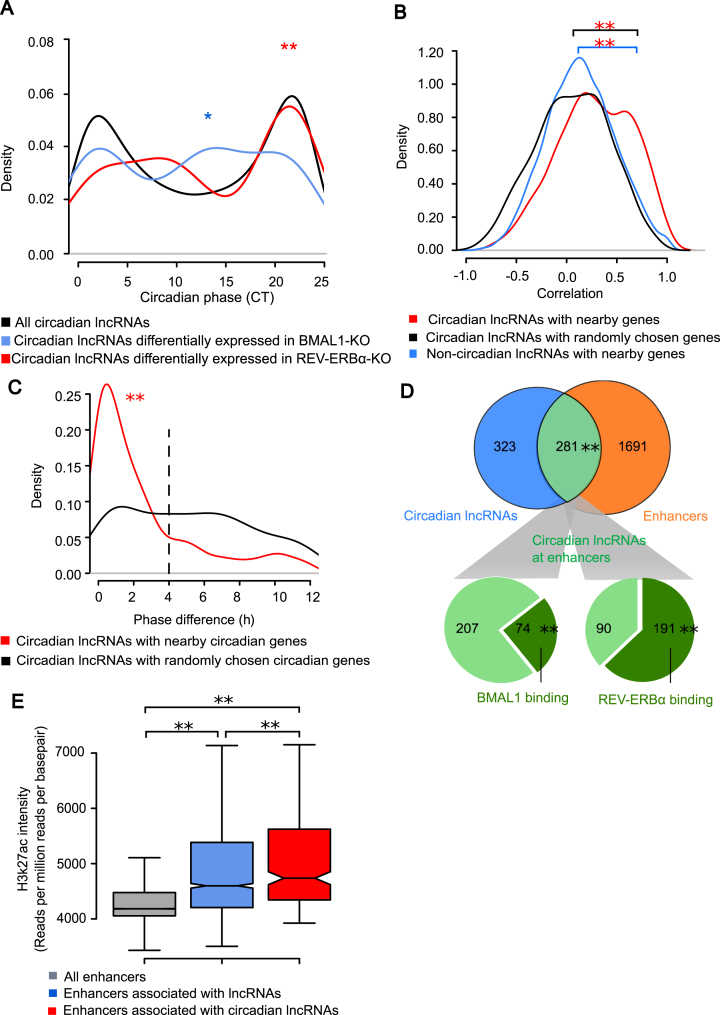
Figure 2.
Circadian lncRNAs were enriched in enhancer regions. (A) Phase distribution of all circadian lncRNAs and those differentially expressed in BMAL1 KO or REV-ERBα KO mice. Phases of circadian lncRNAs differentially expressed in BMAL1 KO and REV-ERBα KO were enriched around CT12 (CT10–CT14, Fisher's Exact Test, *P-value = 0.025) and CT0 (CT21–CT23, Fisher's Exact Test, P-value = 0.0002) respectively, compared with all circadian lncRNAs. (B) Distribution of Pearson's correlation coefficients of lncRNAs with nearby protein coding genes in dataset 2. Comparisons of distribution were performed using KS test, where **P-value = 8.0 × 10−12 (circadian lncRNAs with nearby genes versus circadian lncRNAs with random genes) and **P-value = 3.6 × 10−15 (non-circadian lncRNAs with nearby genes vs. circadian lncRNAs with random genes). (C) Distribution of phase differences between circadian lncRNAs and nearby circadian protein coding genes. Comparison between two distributions of phase differences was performed using KS test, with **P-value = 7.5 × 10−12. (D) Enhancers, BMAL1 binding sites and REV-ERBα binding sites on circadian lncRNA loci. Significance of **P-value < 2.2 × 10−16 was determined by Fisher's Exact Test for the overlap between circadian lncRNAs and enhancers. The binding sites are over-represented on circadian lncRNA loci at enhancer regions compared to those at non-enhancer regions, where significance of **P-value < 2.2 × 10−16 and **P-value < 2.2 × 10−16 were calculated by Fisher's exact test for BMAL1 binding and REV-ERBα binding respectively. (E) Distribution of H3K27ac ChIP intensities on enhancer regions. Comparisons were performed by KS test between pairs, where **P-value < 2.2 × 10−16 (all enhancers vs. enhancers associated with lncRNAs), **P-value < 2.2 × 10−16 (all enhancers vs. enhancers associated with circadian lncRNAs) and **P-value = 0.0017 (enhancers associated with lncRNAs vs. enhancers associated with circadian lncRNAs) suggested they were all significantly different.