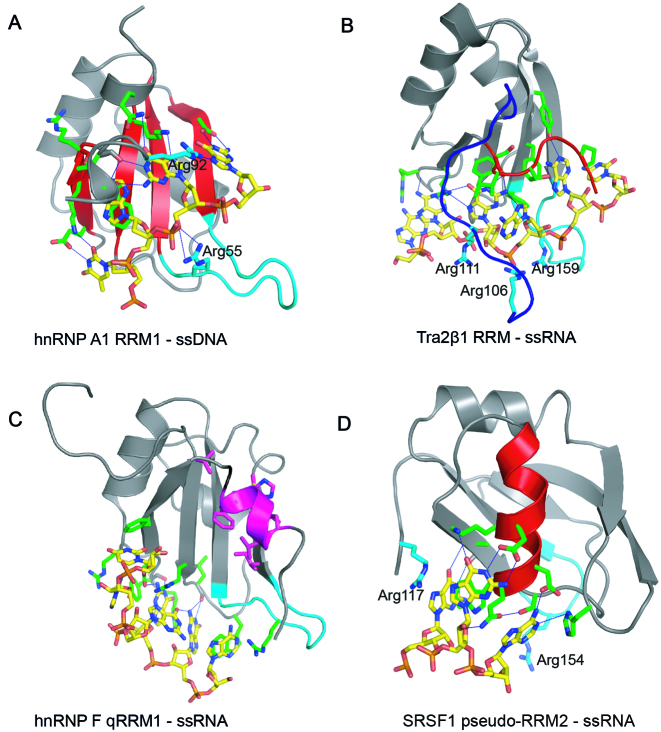
Figure 1.
RRM–ssRNA/ssDNA complexes showing different binding modes. Cartoon representations illustrating the binding of: (A) 5΄–TAGG–3΄ ssDNA to the β-sheet surface (colored red) of hnRNP A1 RRM1 (PDB ID: 2UP1); recognition of –UAGG—RNA sequence is the same. (B) 5΄–AAGAAC–3΄ RNA to the β-sheet surface of Tra2β1 RRM with the involvement of the N- (blue) and C-terminal segments (red) (PDB ID: 2KXN). (C) 5΄–AGGGAU–3΄ RNA to the loop region of hnRNP F qRRM1 (PDB ID: 2KFY); the AGG*G*AU RNA sequence modified by 7-deazaguanine is recognized in the same manner. Residues participating in the hydrophobic core are highlighted in magenta (D) 5΄–AAGGAC–3΄ RNA to α-helix-1 (colored red) of SRSF1 RRM2 (PDB ID: 2M8D). All pictures were created in PYMOL. The proteins and the ssRNA/ssDNA oligonucleotides are colored in gray and yellow respectively. Amino acid side chain contacting the RNA are shown as sticks and colored green. Hydrogen bonds are shown as blue discontinuous lines. Arginine residues involved in ionic bonds are shown as sticks and labeled. The β2/β3 loop region is colored in cyan.