Figure 3.
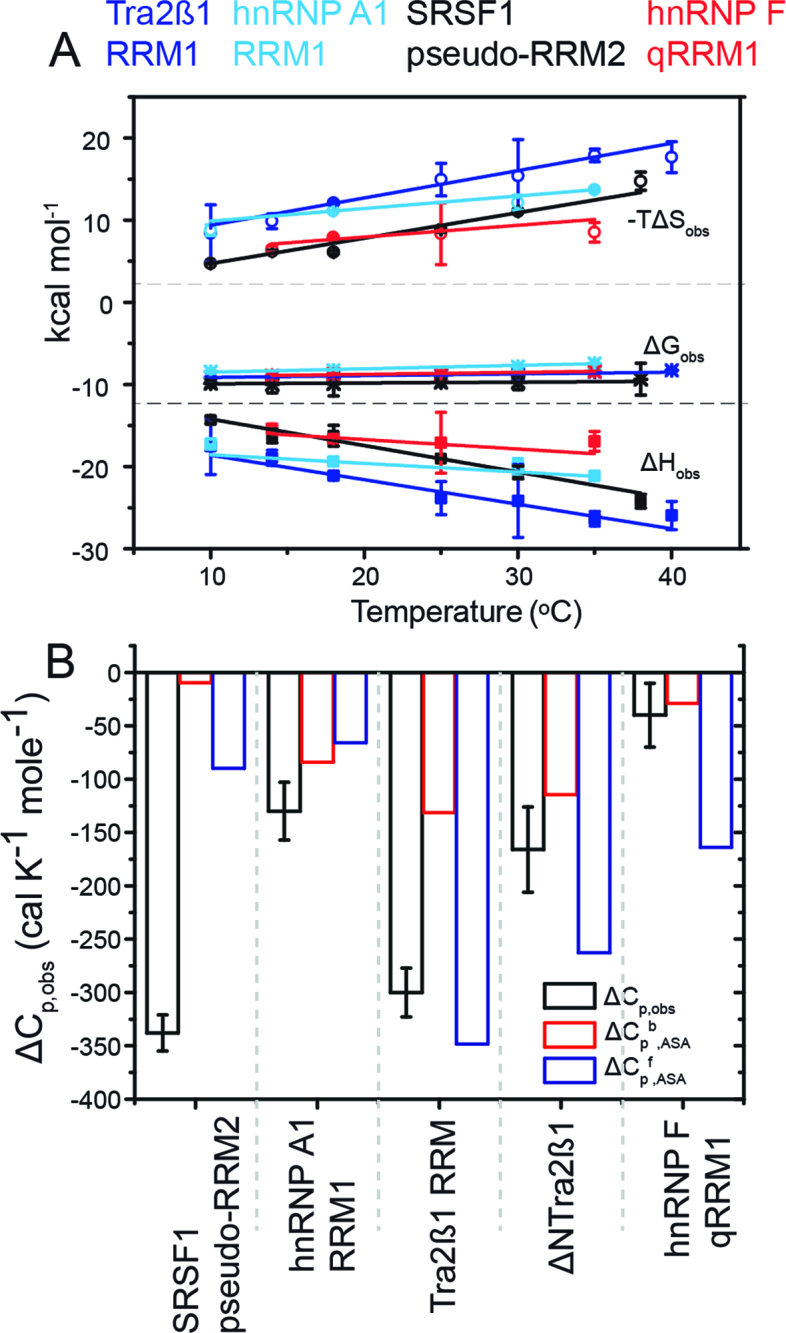
Enthalpy is the general force driving RRM–ssRNA association. Dissection of temperature-dependence of enthalpy change (heat capacity) reveals differential hydration effects, conformational transitions and dynamics upon binding RNA. (A) Plots of the observed molar enthalpy (ΔHobs), molar free energy (ΔGobs) and molar entropy of association (-TΔSobs) determined directly by ITC upon injecting 100–250 μM of RRMs into 5–15 μM cognate RNAs at various temperatures. The thermodynamic parameters were extracted upon fitting Equation (2) to the data shown in Figure 2B. (B) Dissection of heat capacity effects. The experimental observed ΔCp,obs ( = ΔΔHobs/ΔT) is shown in black. The errors are standard deviations of the mean determined by Jackknife analyses of the temperature-dependence of the molar enethalpy. Expected ΔCp was calculated by NACCESS (probe radius and slice-width of 1.4 and 0.05 Å, respectively) based on the bound conformations of the binding partners (ΔCpb,ASA; red) and structures of the free components (ΔCpf,ASA; blue). The structures used in semi-empirical calculations are: hnRNP F qRRM1 (PDB ID: 2HGL, 2KFY), Tra2β1 RRM (PDB ID: 2RRB, 2KXN), hnRNP A1 RRM1 (PDB ID: 1L3K, 2UP1) and SRSF1 pseudo-RRM2 (PDB ID: 2O3D, 2M8D). Models of flexible free RNA oligomers and N- and C-terminal protein segments were modeled using the ff03 force-field in Amber9.
