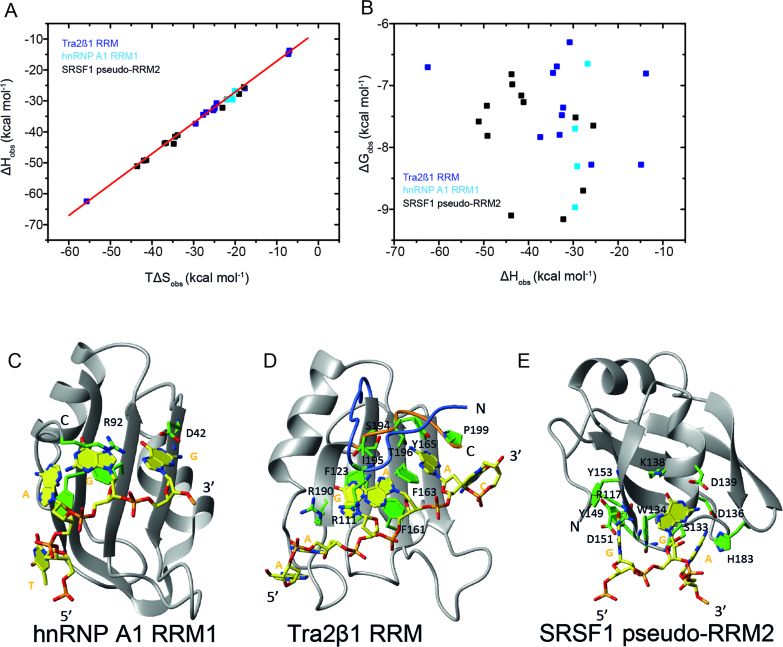
Figure 4.
Networks of weak interactions regulate complex stability in all complexes independent of the mode employed to bind RNA. Plots show the thermodynamic parameters describing the formation Tra2β1 RRM1/5΄-UGAAGAAC-3΄, SRSF1 pseudo-RRM2/5΄-AAGGAC-3΄ or hnRNP A1 RRM1/5΄-UUAGGUC-3΄ protein/RNA complexes determined by ITC titrations after substitution of various amino acids. (A) ΔHobs plotted against TΔSobs (B) ΔGobs plotted against ΔHobs. The experimental temperatures were 313 K (Tra2β1 RRM and SRSF1 pseudo-RRM2) and 303 K (hnRNP A1 RRM1). Protein/RNA titrations of SRSF1 pseudo-RRM2 and Tra2β1 RRM were conducted under the same solution conditions as in Figure 2. Data on hnRNP A1 RRM1 were collected in the presence of 10 mM NaH2PO4, 2 mM β-mercaptoethanol at pH 6.5. Overview of the solution structures of hnRNP A1 bound to the TAGG sequence (C), Tra2β1 RRM bound to the AAGAAC RNA (D), and SRSF1 pseudo-RRM2 bound to the GGA sequence (E). Structure of the complex is shown in ribbon (protein backbone) and stick (RNA) representation. The protein backbone is shown in gray and heavy atoms are shown in orange (P atoms), yellow (C atoms for RNA), green (C atoms for protein), red (O atoms) and blue (N atoms).