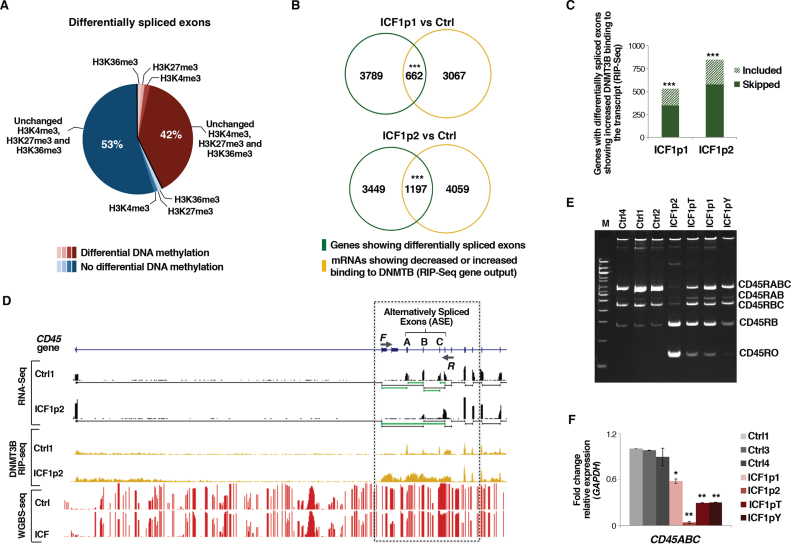
Figure 4.
DNMT3B dysfunction affects alternative exon splicing in ICF1 cells. (A) Pie chart showing the percentage of differentially spliced exonic parts with differentially methylated CpGs and/or H3K4me3, H3K27me3 and H3K36me3 marks changes (statistical analysis was performed using one-tail hypergeometric test, which shows no significant enrichment for these epigenetic modification at differentially included exons); (B) Venn diagram showing the overlap between genes with differentially spliced exons and genes with decreased or increased DNMT3B binding to their transcripts (RIP-Seq) (P-values << 10−16 were calculated using one-tail hypergeometric test); (C) Histogram showing the number of genes with differentially spliced exons and increased DNMT3B binding to the transcript (normalized to IgG fraction; RIP-Seq). Gain of DNMT3B binding significantly associates with skipping or inclusion of exons (P-values << 10−16 were calculated using one-tail hypergeometric test); (D) UCSC genome browser screenshots of PTPRC (CD45) gene showing exon 4–5–6 skipping (RNA-Seq) and CpG methylation status (WGBS); (E) Semi-quantitative PCR amplification of CD45 splicing isoforms in controls and ICF1 samples. Primers used are F and R as shown in D; (F) expression level (qPCR) of CD45RABC using isoform-specific oligonucleotides. *P-adj <0.05; **P-adj <0.005; ***P-adj <0.0005.