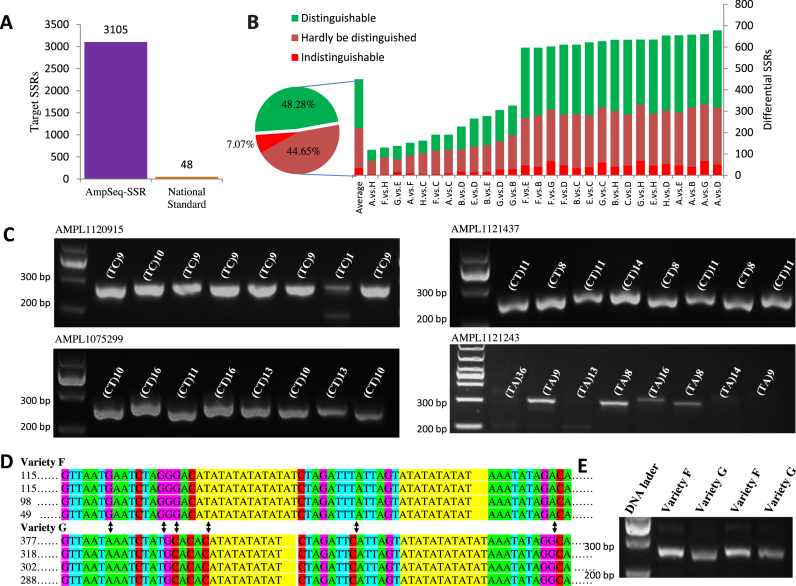
Figure 2.
Highly discriminative fingerprints discovered by AmpSeq-SSR. (A) AmpSeq-SSR employed as many as 3105 target SSRs for rice fingerprint construction, which is ∼65 times of the 48 SSRs listed in rice National Standard. (B) AmpSeq-SSR could detect on average 449.71 differential SSR pairs between any two varieties. Among them, 48.28, 44.65 and 7.07% had amplicon length differences over 5, 1–5 and 0 bp, respectively, and therefore, are respectively distinguishable, hardly distinguished and indistinguishable on electropherograms. (C) An example of SSR with differential SSR genotypes but the same amplicon length. The SSR sequences are shown by letters in yellow background and the nucleotide substitution variations are shown by black arrows. The numbers of sequencing reads are on the left of the sequencing reads. (D) The SSRs in (C) show no difference on agarose gel electropherograms even though they have distinct SSR genotypes. (E) Examples of SSR amplicons on agarose gel electropherograms. On each electropherogram, from left to right are rice varieties A–H marked by their SSR genotypes.