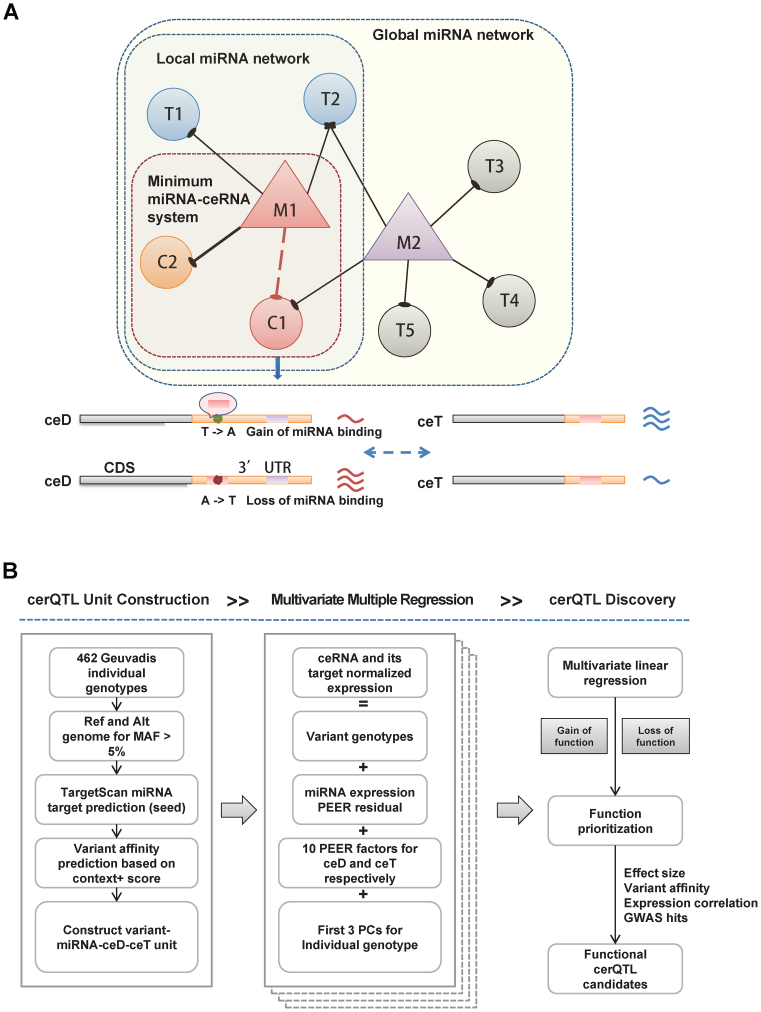
Figure 1.
The schematic diagram of cerQTL detection for genetic variants affecting ceRNA regulation. (A) The model will investigate the variants effect of each ceD-ceT pair in local and unique miRNA-centered regulatory network (minimum miRNA-ceRNA system). If a mutation creates a gain-of-function MRE in the 3΄UTR of an mRNA (C1) targeted specifically by a miRNA (M1), C1 will be treated as a ceD and biologically participates in local network centered on M1. The original targets (including C2, T1 and T2) of M1 will naturally become ceRNA targets (ceTs) of C1. Alternatively, if a loss-of-function mutation erases an MRE of C1 which originally targeted by M1, then C1 will be released from M1-centered regulatory network. Therefore, both scenarios can initiate the perturbation of ceRNA regulation by mutation-driven redistribution of associated miRNA and ceRNAs. Ideally, assuming the concentration of miRNA is relatively stable among investigated individuals, one would observe reciprocal and coordinated expression trend between C1 and each of its ceTs (such as C2) under additive genetic effect. M: miRNA; T: target; C: investigated ceRNA; ceD: ceRNA driver; ceT: ceRNA target. (B) The pipeline first constructs variant-miRNA-ceD-ceT unit according to Geuvaids individual genotypes and TargetScan prediction. For each variant-miRNA-ceD-ceT unit, we test the genotype effect of ceRNA regulation using a multivariate multiple linear regression with two responses (ceD and ceT) by incorporating several confounding factors. Also, different function evidences area applied to prioritize the detected cerQTLs.