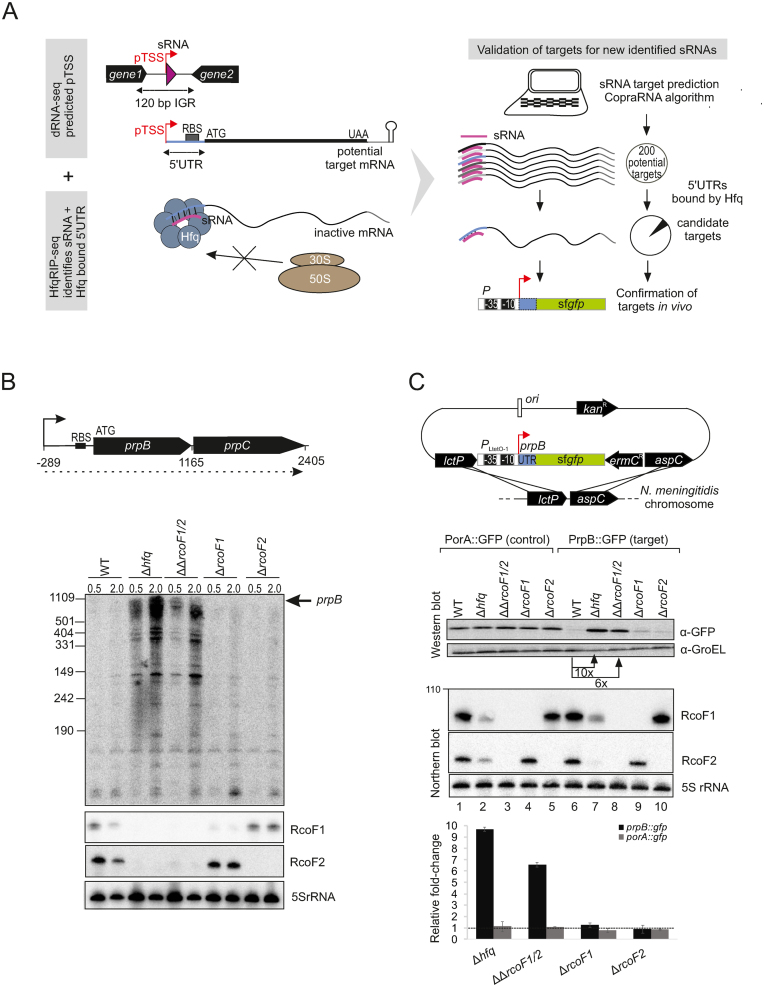
Figure 7.
In vivo validation of prpB regulation by the two sRNAs RcoF1 and RcoF2. (A) (Left) Schematic representation for combining data gained from dRNA-seq and Hfq RIP-seq analysis to uncover and validate targets for newly identified sRNAs. dRNA-seq enriches for primary transcriptional start sides (pTSS) as indicated by red arrows and therewith uncovers sRNAs (pink triangles) located in IGR and other transcriptome features like 5΄ UTRs of mRNAs colored in blue. Hfq RIP-seq allows for the identification of sRNAs and 5΄ UTRs as targets of Hfq. 5΄ UTRs are target sites for Hfq-dependent sRNA regulation that leads to inhibition of ribosome binding, resulting in reduced translation. (Right) Schematic workflow for target prediction by the copra algorithm and validation via a superfolder gfp (sfgfp) reporter gene fusion, exemplified by the two newly identified sRNAs RcoF1 and RcoF2 and their target prpB. Hfq-bound 5΄ UTRs of mRNAs identified by Hfq-RIPseq (colored in blue) serve to further screen potential mRNA targets predicted for new sRNAs identified by dRNA-seq (colored in pink). (B) (top) The 1165-nt-long prpB transcript is part of the dicistronic mRNA prpB-prpC (dotted line) encoding a methylcitrate lyase and methylcitrate synthase, respectively. The TSS is indicated by arrows and the ribosome binding site (RBS) by a black bar. Numbers indicate the distance to the TSS. (bottom) The expression of prpB was analyzed during two growth phases in WT, Δhfq, ΔΔrcof1/2 double deletion mutants as well as Δrcof1 and Δrcof2 single deletion mutants at indicated optical densities (OD600) using a 32P-UTP labeled riboprobe. RcoF1 and RcoF2 were detected with JVO13282 and JVO13283. 5S rRNA served as loading control. (C) (top) Schematic illustration of the integrational vector pGCC2 for construction of translational superfolder gfp (sfgfp) fusions that were inserted into the lctP and aspC locus of N. meningitidis 8013 (127). The prpB 5΄ UTR and the first 15 amino acids of the prpB coding region (gray box) were fused to sfgfp reporter gene (green box) that is transcribed from the constitutive PLtetO-1 promoter as indicated. Individual elements are not drawn to scale. A sfgfp reporter fusion expressing the 5΄ UTR and the first 15 amino acids of the porA gene served as control. (bottom) N. meningitidis 8013 WT, Δhfq, ΔΔrcof1/2 double deletion mutants as well as Δrcof1 and Δrcof2 single deletion mutants expressing either the target-gfp or the control-gfp fusions were grown to mid logarithmic phase and RNA and protein samples were analyzed by northern blot and western blot. RcoF1 was detected with JVO-13282 and RcoF2 was detected with JVO-13283. 5S rRNA served as loading control. Whole cell protein fractions (OD600 = 0.01 for western blot) were detected with mouse anti-Gfp antiserum. GroEL served as control. Relative fold expression changes of prpB::gfp and porA::gfp (control) fusions in Δhfq, of both sRNAs RcoF1 and RcoF2 or of each sRNA alone determined by western blot analysis for Gfp in comparison with the respective WT backgrounds are represented in the bar plot. Error bars indicate the SDs among three biological replicates.