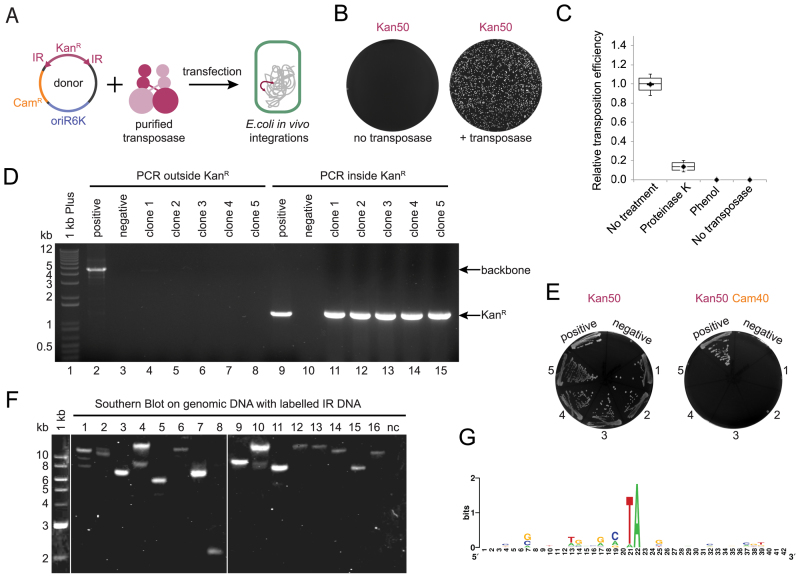
Figure 2.
In vivo transposition in prokaryotic cells. (A) Scheme of the experimental method. The donor plasmid DNA, carrying a gene of interest (kanamycin resistance cassette, KanR) flanked with (IRs), contains a conditional origin of replication, oriR6K, which prevents replication in the recipient strain. Donor plasmid DNA and purified recombinant transposase (Mos1 or Mboumar-9) are co-transfected into bacterial cells, resulting in integration of the kanamycin cassette into the genomic DNA. (B) Kanamycin resistant colonies were obtained only if purified transposase was included in the transfection reaction. White colonies on dark background. (C) Relative efficiency of transposition observed after treatment of the reactions, prior to transfection, with proteinase K, phenol, no treatment and the control (no transposase added). Two technical repeats were performed for two biological repeats. (D) Agarose gel of the products of colony polymerase chain reaction (PCR) to detect the presence of the donor plasmid in the kanamycin resistant colonies. Clone 1 (Lane 4) has traces of the donor plasmid backbone detected. Positive control—a colony of Escherichia coli S17 λ pir carrying donor plasmid; negative control—a colony of the recipient strain E. coli DH10B. (E) Five analyzed clones are resistant to kanamycin, but sensitive to chloramphenicol—the plasmid backbone resistance. Controls as in (D). (F) Southern Blotting analysis of the digested genomic DNA from the kanamycin resistant clones hybridized with fluorescently labeled IR DNA. Negative control: genomic DNA of the recipient strain E. coli DH10B. (G) WebLogo alignment of 40 bp around the TA target nucleotides duplication of the 14 integration sites by Mos1 in the bacterial genome.