Nucleic Acids Research, (2015) 43(18):9051–9064. doi: 10.1093/nar/gkv848
We have re-examined the crystallographic data and carefully re-refined all the three structures reported in the paper. The re-refinement process and results of each structure are as follows:
1. Structure of Smad5-MH1/GC-BRE
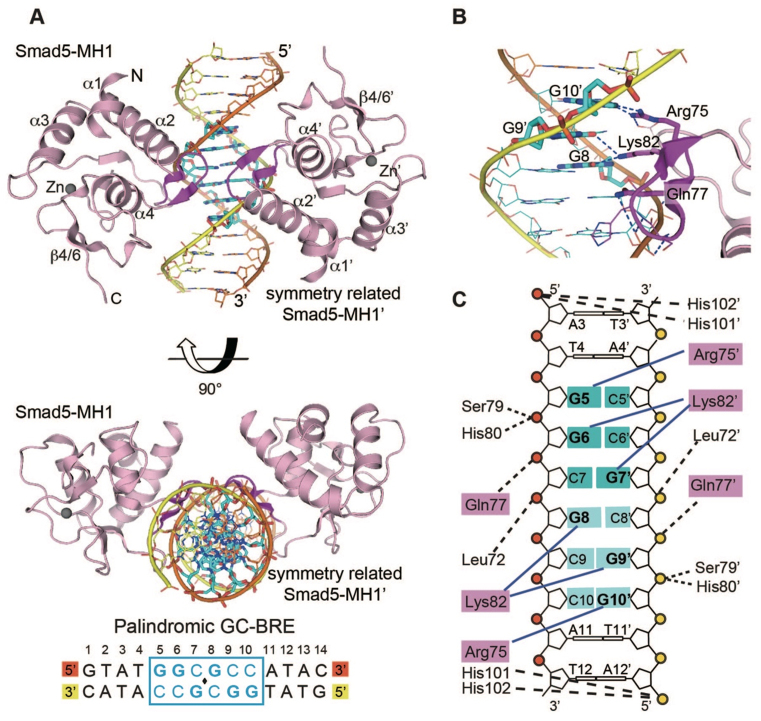
The space group for the crystal structure of Smad5-MH1/GC-BRE was previously assigned as P31. After re-examining the crystallographic data, however, we wish to revise the space group to a higher symmetry one, P6422. As the sequence of GC-BRE DNA is palindromic, the revised structure contains one Smad5-MH1 monomer and a single GC-BRE DNA strand in an asymmetric unit, where the 14 ordered nucleotides form a double helix with a symmetry-related DNA strand. The physiological complex of two Smad5-MH1 monomers and one GC-BRE DNA duplex (the 2:1 Smad5-MH1/GC-BRE complex) is basically the same as the previous Smad-DNA complex of P31 space group, with a Cα RMSD of ∼0.4 Å for 180 aligning residues.
Owing to the limited resolution, not all side chains could be placed with high confidence and the side chains of some residues were removed. Fortunately, residues involved in the protein-DNA interactions are mostly ordered, and the interaction mode of two Smad5-MH1 monomers with the GC-BRE DNA duplex is unchanged. As the space group of this structure is changed, we revise Figure 3 (relabeling the molecules and side chains) and Supplementary Figure S3 (replacing panels A and B).
Figure 3.
Crystal structure of Smad5-MH1 in complex with the GC-BRE sequence. (A) Overall structure of the physiological 2:1 Smad5-MH1/GC-BRE complex. The Smad5 MH1 domains are coloured in light pink, with the β-hairpin highlighted in magenta. The zinc atoms are shown as spheres. The symmetry-related Smad5-MH1 molecule is labled with an apostrope. The template chain and the symmetry-related complimentary chain of the dsDNA are shown in orange and yellow, respectively. The DNA bases of the central GC-BRE site are shown as sticks and coloured in cyan. The DNA sequence used for crystallization is shown below the structure, with the central 6-bp GC-BRE site boxed and highlighted in blue. (B) Detailed interactions between the β-hairpin and the DNA bases. Three conserved β-hairpin residues and the DNA bases involved in specific interactions are highlighted as magenta and cyan sticks, respectively. Hydrogen bonds are represented by blue dashed lines. (C) Schematic drawing of Smad5-MH1 binding to the GC-BRE sequence. Non-interacting nucleotides are omitted. Each copy of the GC-rich motif is shaded in dark cyan and light cyan, respectively. The conserved β-hairpin residues, Arg75, Lys82 and Gln77, are shaded in pink. Interactions between the amino acids and DNA bases are shown as blue solid lines, while the contacts with the DNA phosphates are shown as black dashed lines. Residues and DNA bases in the symmetry-related molecules are indicated by apostropes.
2. Structure of Smad5-MH1/GCRj2
We also re-refined the structure of Smad5-MH1 in complex with the composite Smad binding sequence GCRj2. The entire refinements were performed using phenix.refine, and the torsion-based NCS restraints (with the default setting by phenix.refine) were applied throughout. We have corrected most non-rotameric side chains and removed several side chains because of the poor electron density. To note, the changes in structure were made in regions distant from the protein-DNA interface, and the interaction mode between Smad5-MH1 with either the SBE site or the GCC site described in the paper is unaffected.
3. Structure of Smad5-MH1/SBE
We also supersede PDB 4ZKG (structure of Smad5-MH1 in complexes with a palindromic SBE DNA) with the new coordinate similarly re-refined (correcting non-rotameric side chains, removing side chains of poor density, and applying the NCS restraints throughout the refinement). The revision of structure does not change the scientific conclusions in the paper.
4. Revised Table of crystallographic data collection and refinement statistics
All three re-refined structures have been deposited to PDB to supersede previous entries. The electron density maps are available for download in the PDBe website and the SF-TOOL web server. Please find herewith the revised version of Table 1, which contains the structure statistics for the updated structures.
Table 1. Crystallographic data collection and refinement statistics.
| Smad5-MH1/SBE | Smad5-MH1/GC-BRE | Smad5-MH1/GCRj2 | |
|---|---|---|---|
| Old PDB ID code | 4ZKG | 4ZL2 | 4ZL3 |
| New PDB ID code | 5 × 6G | 5 × 6H | 5 × 6M |
| Data Collectiona | |||
| Space group | P212121 | P6422 | P31 |
| Cell dimensions | |||
| a, b, c (Å) | 71.54, 74.50, 83.74 | 92.87, 92.87, 83.71 | 119.46,119.46, 93.07 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 50.00-3.05 | 50.00-3.10 | 40.00-3.20 |
| (3.10-3.05)b | (3.21-3.10) | (3.31-3.20) | |
| R merge (%) | 7.7 (44.4) | 6.2 (44.9) | 9.5 (55.6) |
| I/σ (I) | 16.1 (4.7) | 25.0 (4.28) | 22.1 (4.5) |
| Completeness (%) | 89.9 (100.0) | 97.4 (100.0) | 98.8 (100.0) |
| Redundancy | 6.5 (7.2) | 7.2 (7.5) | 5.5 (5.7) |
| Refinement | |||
| Resolution (Å) | 34.04-3.05 | 46.43-3.10 | 29.94-3.20 |
| No. of reflections | 7870 | 4079 | 24146 |
| R work/Rfreec (%) | 22.3/25.6 | 20.2/23.1 | 21.1/25.3 |
| No. Atoms | |||
| Protein/DNA | 2571 | 1136 | 5379 |
| Zinc | 2 | 1 | 4 |
| Average B-factors | 90.2 | 96.1 | 125.1 |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.010 | 0.011 | 0.011 |
| Bond Angles (°) | 1.21 | 1.23 | 1.23 |
| Ramachandran analysis (%) | |||
| Favored | 98.8 | 97.3 | 96.4 |
| Additionally allowed | 1.2 | 2.7 | 3.6 |
| Disallowed | 0 | 0 | 0 |
All data sets were collected from a single crystal.
Values in parentheses are for the highest resolution shell.
R free was calculated on a random 5.0% reflections of the data.
We regret the negligence in the structure refinements and apologize to the readers and publisher for the inconvenience. Fortunately, residues involved in the protein-DNA interactions are mostly ordered in all three structures, and the interaction modes of Smad5-MH1 with either the GC site or the SBE site are unchanged. The revision of these structures does not affect the conclusions and discussion in the paper.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.