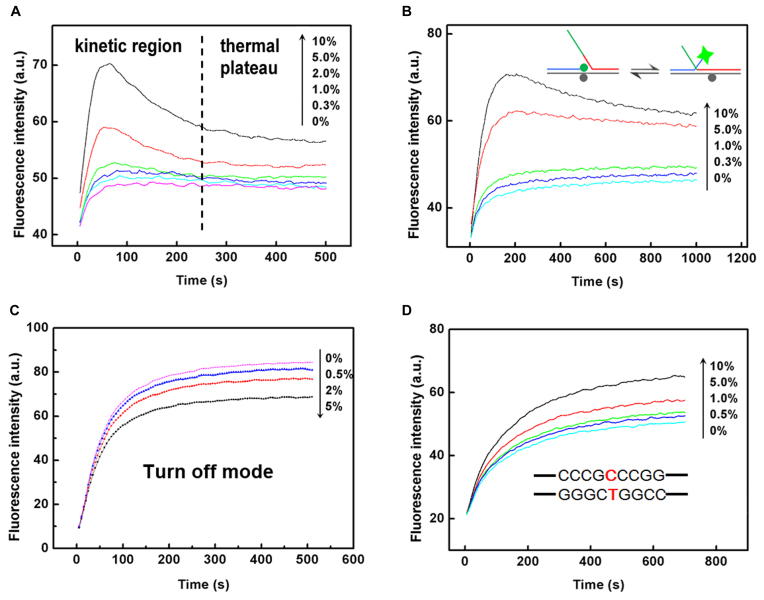
Figure 3.
Fluorescence curves show the capability of the BM probe in discrimination of low-abundance mismatched base pairs in a target strand with different length and surrounding sequences. (A) The fluorescence intensity responses of BM-1 probe toward PM-1 target in the presence of different amount of 1-MM-1 target. (B) The fluorescence intensity responses of BM-1 probe toward PM-1L target in the presence of different amount of 1-MM-1L target. The overhangs in the PM target and 1-MM target are shown in green. (C) Performance of BM-1 probe in a turn-off mode for the discrimination of low-abundance 1-MM-1L-T target by employing PM-1L as the wild-type DNA. (D) The fluorescence intensity responses of BM-2 probe toward PM-2 target at different abundances when the mutation site is surrounded by guanines and cytosines. Such point mutations are difficult for enzyme-dependent probes to identify. We chose an EGFR associated point mutation (NM_0005228.3:c.2573T>G) as our model target here.