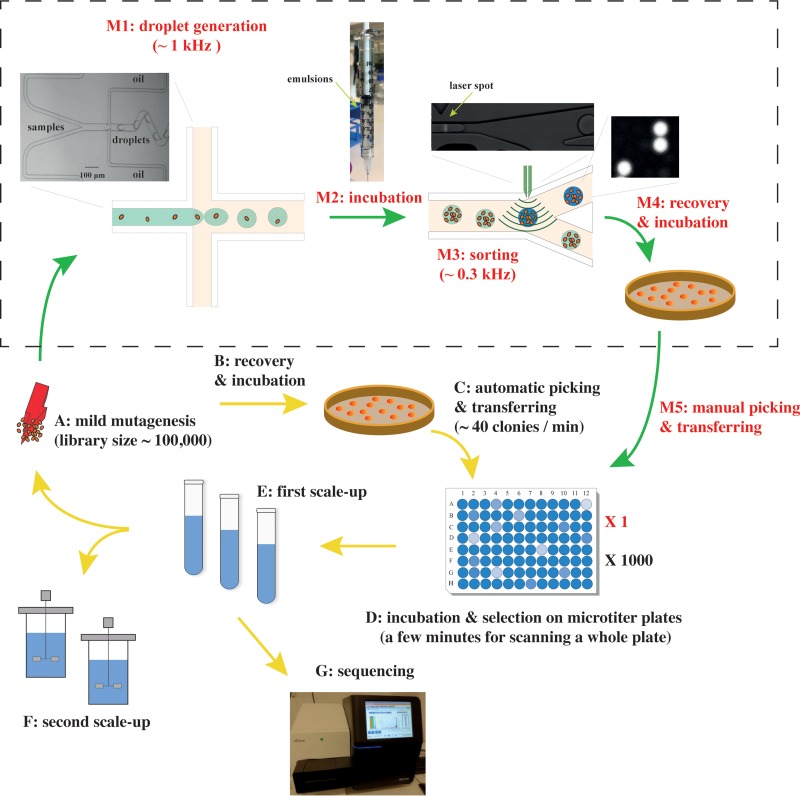
FIG 1 .
Incorporation of the droplet-based microfluidic screening toolbox into the classical strain development process based on random mutagenesis and microplate screening. Steps A to G represent the classical protocol, and the microfluidic toolbox (M1 to M5) is inserted between steps A and D. (A) Mutagenesis is induced by physical and chemical mutagens, such as UV and EMS. Mutation frequency is controlled by either the dosage used or the reaction time. (B) Prior to being selected by robots, well-separated colonies should have formed on agar plates. (C) Assuming that the automatic selection and transfer rate is 40 clones/minute (5), the process is completed in approximately 40 h for a library containing 100,000 clones. (D) One thousand 96-well microplates are required in the protocol that does not use the microfluidic toolbox. (M1 and M3) notably, if a Poisson distribution λ of 0.5 (60% empty droplets) is applied, the true rates for the generation and sorting of cell-loaded droplets are 60% lower. (M2) The conditions for incubation are the same as in steps B and M4. In this study, a 24-h incubation at 30°C is typically required for L. lactis. (M4) Cells in droplets are released by the addition of PFOH (1H,1H,2H,2H-perfluorooctan-1-ol). (M5) Due to the efficient enrichment (2,000-fold) using microfluidic screening, fewer than 90 clones are selected for the secondary screening in 96-well microplates.