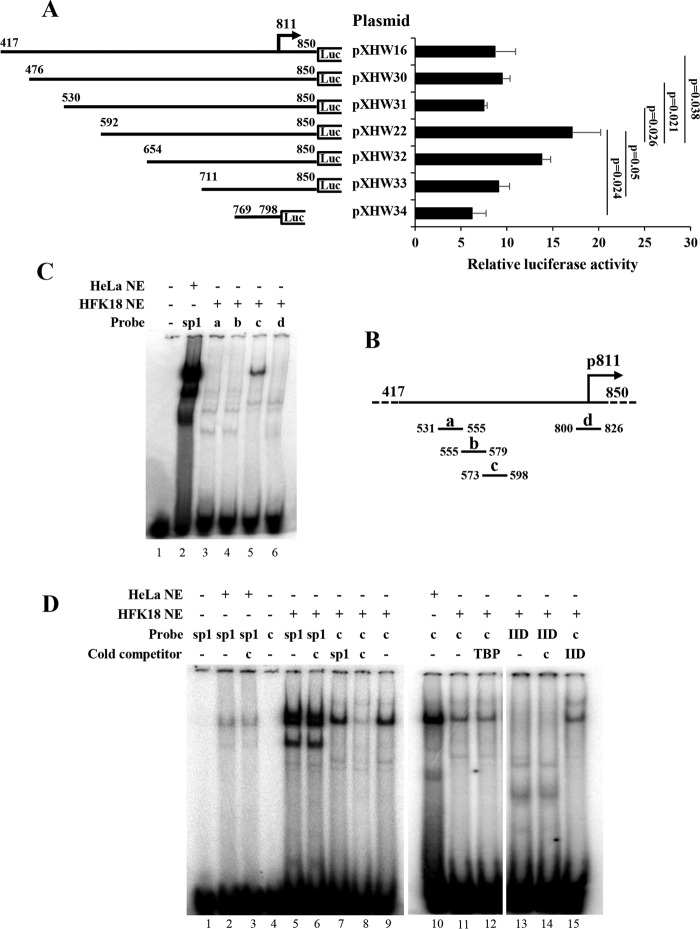
FIG 4 .
Identification of a transcriptional repressor element that affects P811 promoter activity and binds cellular proteins. (A) Schematic diagrams and their promoter activities for individual plasmids derived from pXHW16 with indicated deletions of the promoter region (nt 417 to nt 850 in the HPV18 genome) inserted upstream of a firefly luciferase (Luc) gene. The numbers above the lines represent nucleotide positions of the first and last nucleotides of the insertion in the HPV18 genome. The HPV18 late TSS at nt position 811 is indicated by an arrow. HFK18 cells in the presence of 2 mM calcium were cotransfected for 48 h with the individual plasmids along with Renilla luciferase plasmid pRL-TS. The supernatant of the cell lysates was examined for dual luciferase activities, and the relative promoter activity levels were calculated as described for Fig. 2A. (B) Diagrams of the nucleotide positions of four synthetic, double-stranded DNA oligonucleotide probes (a to d) used for electrophoretic mobility shift assays (EMSA). (C) Probe c, derived from nt 573 to nt 598 in the HPV18 genome, interacts with a cellular protein(s) from HFK18 cells. Probes were labeled with 32P, incubated with nuclear extract (NE) from HeLa or HFK18 cells, and then examined by EMSA. A Sp1 consensus oligonucleotide probe was used as a positive control. Protein-DNA complexes were resolved on a 4% native polyacrylamide gel. (D) Verification of the cellular proteins interacting with the repressor element by competitive gel shift assays. 32P-labeled probe c and a 32P-labeled Sp1 or TFIID (IID) oligonucleotide were incubated with NE prepared from HeLa or HFK18 cells in the presence or absence of an indicated cold competitor probe c, Sp1, TBP, or IID consensus oligonucleotide. Protein-DNA complexes were resolved on a 4% native polyacrylamide gel.