FIGURE 2.
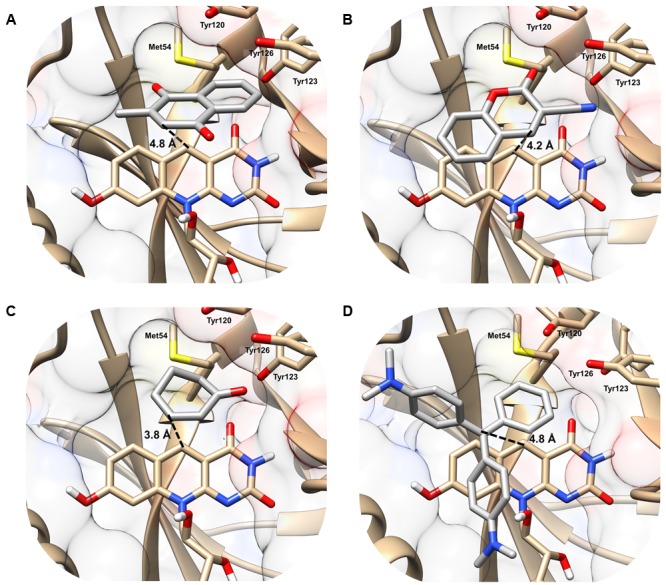
Structural basis of substrate activation by F420H2-dependent reductases. The secondary structure and surface rendering of the cofactor- and substrate-binding site of MSMEG_2027 are shown based on the 1.5 Å resolution crystal structure (PDB: 4Y9I) (Ahmed et al., 2015) of the enzyme. The structures are computationally docked with (A) menadione, (B) 3-cyanocoumarin, (C) 2-cyclohexen-1-one, and (D) malachite green. The distance between the proposed hydride donor (C5 of F420H-) and hydride acceptor (electrophilic carbon of the substrate) are shown. Residues within 5 Å of the substrate are shown. Docking results with the more specific F420H2-dependent reductase MSMEG_6526 are shown in Supplementary Figure S2 and are compared with MSMEG_2027 in Supplementary Table S4.
