Fig. 1.
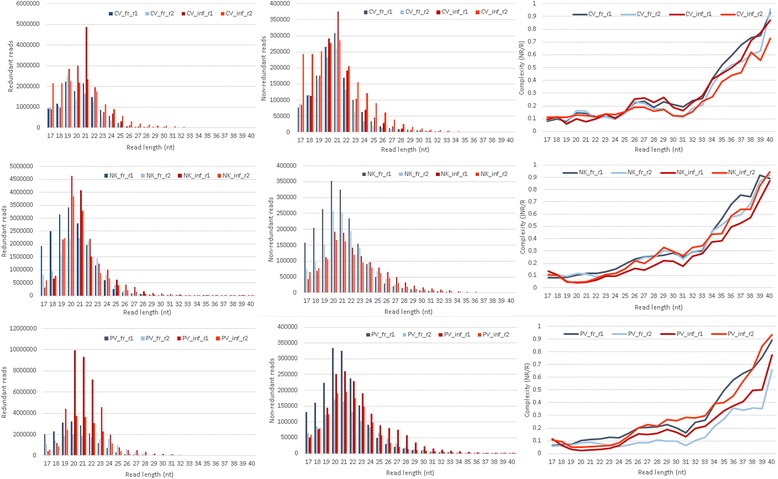
Size class (redundant, R and nonredundant, NR) and complexity (C) distributions for all samples after adapter removal and before the alignment to the reference genomes for the virus-free and virus-infected samples. The CV, NK and PV correspond to Aspergillus fumigatus chrysovirus (AfuCV), a strain of Aspergillus fumigatus tetramycovirus-1 (AfuTmV-1) and Aspergillus fumigatus partitivirus-1 (AfuPV-1), respectively. Histograms show the size class distribution of all (redundant; R) and unique (non-redundant; NR) reads and also the complexity (C) of the samples. On the x-axis we represent the size classes from 17 to 40 nucleotides (nt). On the y-axis we represent the overall abundance (for the R distribution), the frequency of reads in each size class (for the NR distribution) and the complexity (for the complexity distribution). The complexity, calculated as the ratio of non-redundant to redundant reads, varies between 0 and 1 [37]. Low complexity values (close to 0) indicate a small group of highly abundant sequences, values close to 1 indicate the presence of a large group of sequences with low abundance. (CV_fr_r1: AfuCV free isolate replicate-1; CV_fr_r2: AfuCV free isolate replicate-2; CV_inf_r1: AfuCV infected isolate replicate-1; CV_inf_r2: AfuCV infected isolate replicate-2; NK_fr_r1: AfuTmV-1 free isolate replicate-1; NK_fr_r2: AfuTmV-1 free isolate replicate-2; NK_inf_r1: AfuTmV-1 infected isolate replicate-1; NK_inf_r2: AfuTmV-1 infected isolate replicate-2; PV_fr_r1: AfuPV-1 free isolate replicate-1; PV_fr_r2: AfuPV-1 free isolate replicate-2; PV_inf_r1: AfuPV-1 infected isolate replicate-1; PV_inf_r2: AfuPV-1 infected isolate replicate-2)
