Fig. 4.
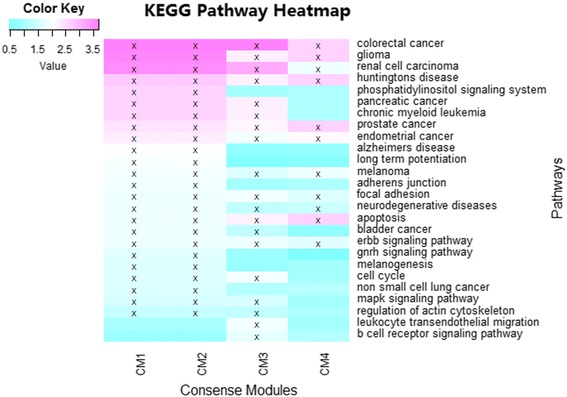
Functional annotation of the four identified consensus modules (CM1-CM4) using KEGG pathways. The four consensus modules were treated as four gene sets, and went through pathway enrichment analysis based on the KEGG pathway database. The enrichment results at a nominal statistical threshold of adjusted p-value < 0.05 are shown. -log10(p-value) values are color-mapped and displayed in the heat map. Heat map blocks labeled with “x” reach the nominal significance level of adjusted p-value < 0.05. Only top enrichment findings are included in the heat map, and so each row (pathway) has at least one “x” block
