Fig. 3.
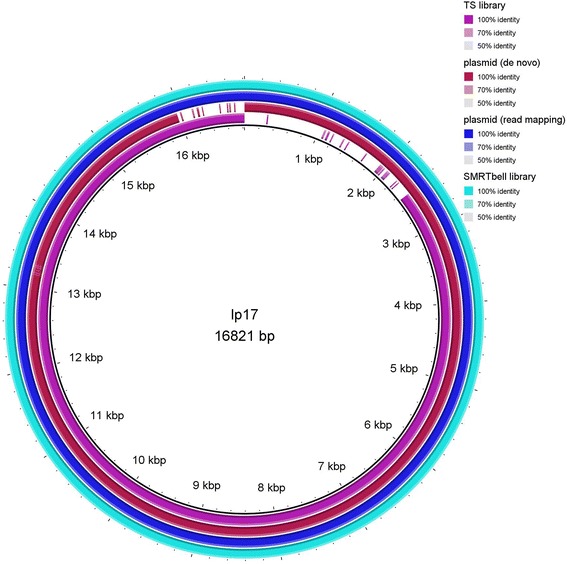
Visualization of assemblies of lp17 in PAbe aligned to the reference B31-GB using BRIG. Total genomic DNA was used for TS library preparation (purple color, labelled TS library). A spurious gap in the region from 0 to 2.5 kb is visible. Using enriched plasmid DNA for NX library construction improved the assembly and a smaller gap at 16–17 kb is visible (red color, labelled plasmid (de novo)). Not surprinsingly, read mapping of Illumina reads on B31-GB using enriched plasmid DNA for library construction showed complete coverage suggesting that reads for the complete plasmid existed but were not assembled de novo (blue color, labelled plasmid readmapping). De novo assemblies of Pacific Bioscience SMRT sequences showed complete coverage of lp17 confirming that the complete plasmid was present in PAbe and that the gaps presenting in de novo assemblies of short reads were artefacts either of library construction or short read assembly (torquois color, labelled SMRT Bell library)
