Fig. 4.
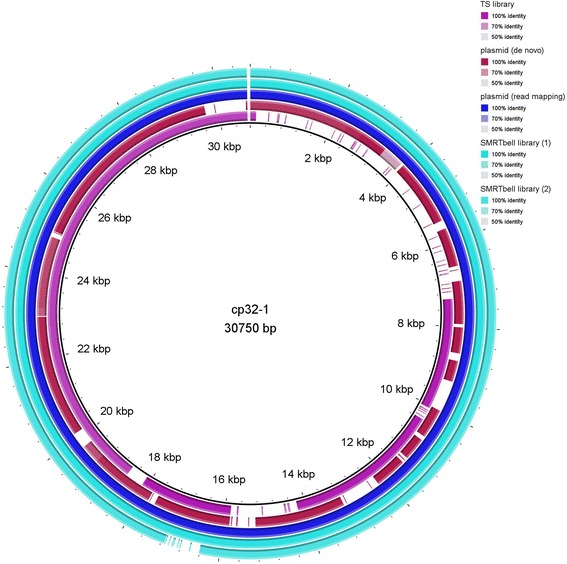
Visualization of assemblies of cp32-1 of B31-NRZ aligned to the reference B31-GB. De novo assembly of total genomic DNA sequenced from a TS library preparation (purple color, labelled TS library). The alignment shows several gaps covering only three quarter of the plasmid. Using enriched plasmid DNA for NX library construction followed by de novo assembly in the CLC Genomics workbench (red color, plasmid (de novo)), the size of gaps was reduced. Read mapping of Illumina reads on B31-GB using enriched plasmid DNA for NX library construction (blue color, plasmid (read mapping)) produced a gapless alignment of reads to the reference. De novo assemblies of Pacific Bioscience SMRT sequences (torquois color, SMRT Bell library) showed complete coverage of the cp32-1 plasmid. The image shows the larger size of the SMRT assembly with a gap appearing around 17 kb which likely reflect the differences observed between B31-NRZ and B31-GB. Different shades of coloration in one panel indicate different identities between aligned sequences. Regions with lighter shades correspond to less sequence similarity (see region app. 3.5 kbp to 4.0 kbp in de novo assembly using enriched plasmids)
