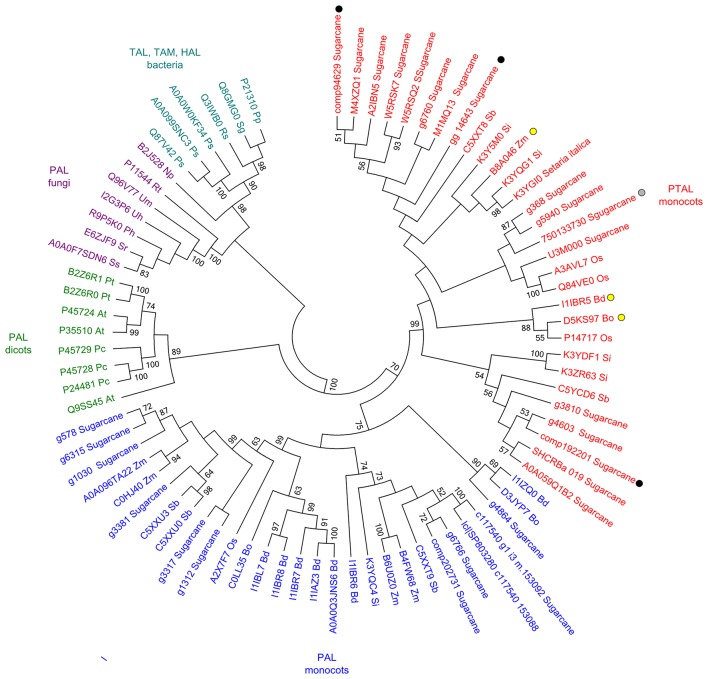
Figure 5.
Sequence analysis of PAL and PTAL proteins. Phylogenetic tree of PTAL and PAL in plants and fungi, and TAL, tyrosine ammonia-mutase (TAM) and histidine ammonia-lyase (HAL) in bacteria. Multiple sequence alignments obtained in ClustalW implemented in MEGA 6.06 Software (Tamura et al., 2013) with default settings. Maximum-likelihood algorithm was used to build the phylogenetic tree with the following settings: JTT model, 1,000 replicates of bootstrap analyses, with the best network-network interface (NNI) topology search. Protein sequences were obtained from Uniprot and GenBank (Supporting Information File S1). Black circles: sugarcane genes encoding the enzymes up-regulated in plants after whip development (Schaker et al., 2016); Gray circle: sugarcane protein identified exclusively in sugarcane plants after whip development (Barnabas et al., 2016); Yellow circles: previously reported PTAL proteins (Barros et al., 2016; Maeda, 2016).