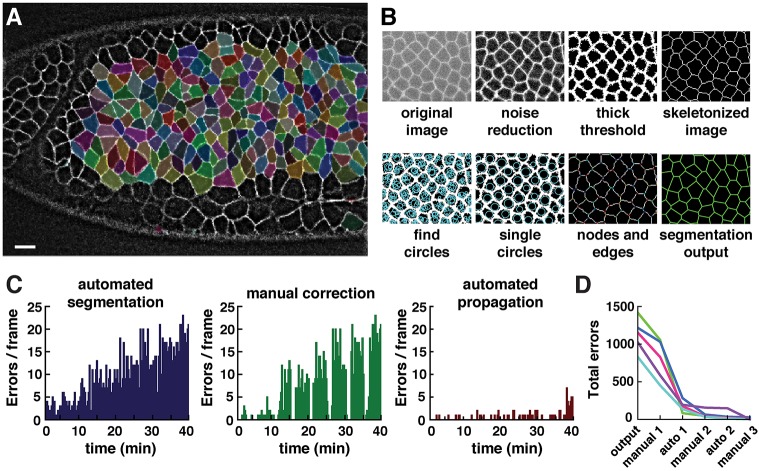
Fig. 2.
Automated and semi-automated tools in SEGGA enable rapid and accurate cell segmentation. (A) Segmented cells in a wild-type Drosophila embryo during axis elongation (white, Resille:GFP). Cell shapes are well represented by a polygonal lattice (cells highlighted in random colors). Anterior left, ventral down. Scale bar: 10 µm. (B) Overview of image segmentation steps in SEGGA. (C) Errors in the initial segmentation output (left) were manually corrected in every twentieth frame (middle) and corrections were automatically propagated to other time points (right). Images were acquired every 15 s; n=average of 390 cells/image. (D) In general, three rounds of manual correction and two rounds of automated propagation produced 100% accurate tracking for movies of Drosophila axis elongation. n=5 movies; each line represents one movie.