Figure 5. Complex models of HSD evolutionary history.
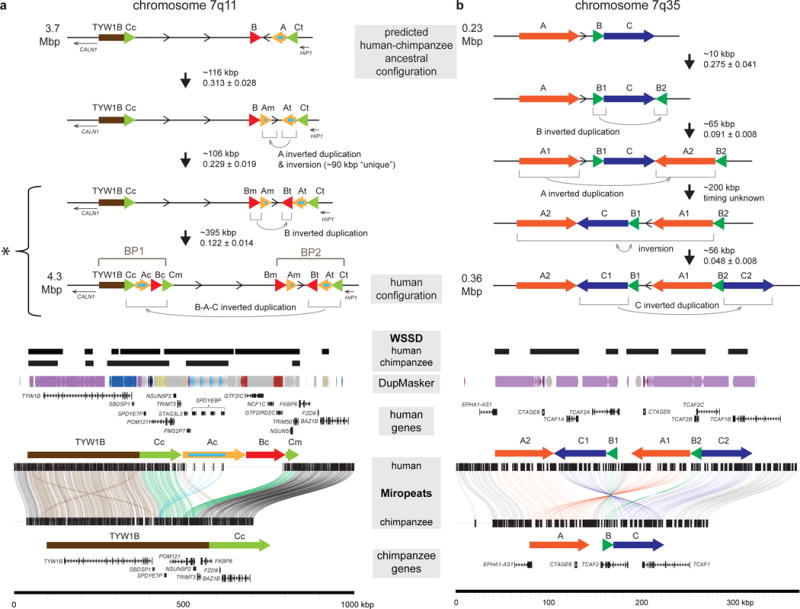
BACs tiling across human chromosome (a) 7q11 and (b) 7q35 regions were sequenced and assembled (representing human and additional great apes) and supercontigs were created. Estimates of sizes and evolutionary timing (human–chimpanzee distance; Supplementary Table 8) of events are denoted between each predicted intermediate genomic structure. SD organization is depicted as colored arrows across the 7q11 (SDs annotated with subscripts representing relative positions including centromeric (c), middle (m), and telomeric (t) as previously defined41) and 7q35 regions. The orientations of intervening regions are shown with arrows. Models of the predicted evolutionary histories of the HSDs at all loci are depicted starting with the predicted human–chimpanzee common ancestor to the most common haplotype present in modern humans. A Miropeats comparison of the human and chimpanzee contigs shows the pairwise differences between the orthologous regions. Lines connect stretches of homologous regions based on a chosen threshold (s), defined as the number of matching bases minus the number of mismatching bases (s = 500 for a, s = 1000 for b) and match the arrow colors when they connect SD blocks. Additional annotations include whole-genome shotgun sequence detection (WSSD) in human and chimpanzee, indicating duplicated regions identified by sequence read depth56, DupMasker79, and genes.
