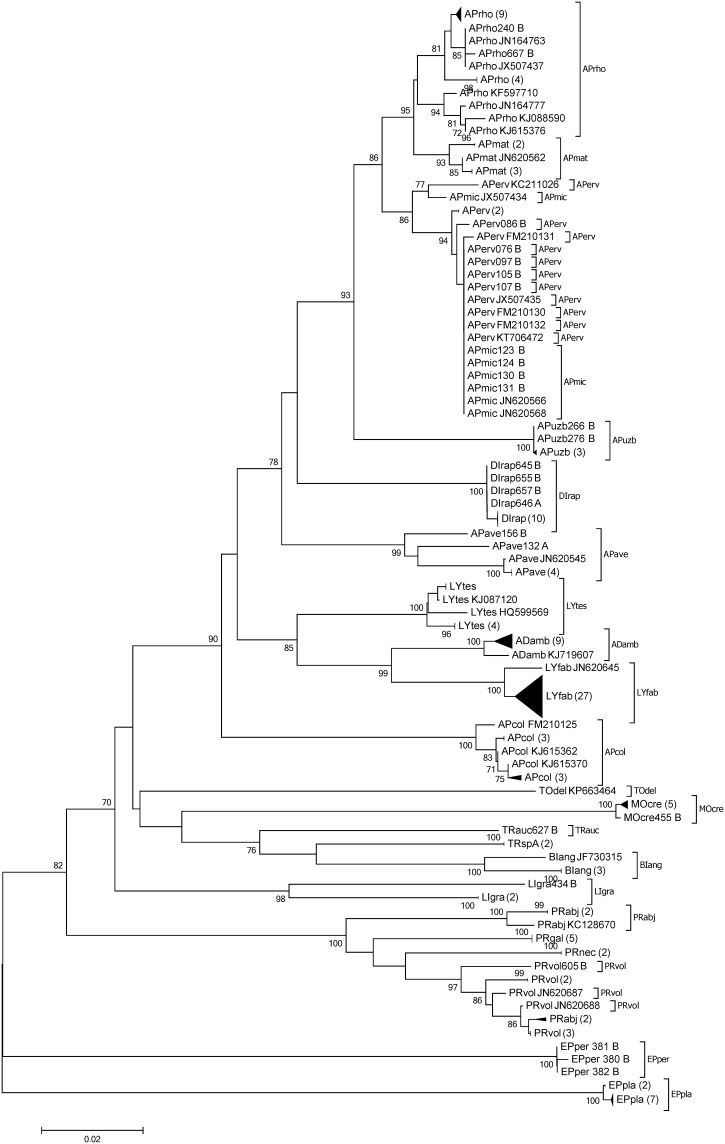
Fig 4. Neighbour joining tree of parasitoid species within the Aphidiinae based on sequences of the 5’-region of the cytochrome c oxidase I gene (COI).
Bootstrap values (≥ 70%) are indicated on branches. Identical sequences and subtrees with all bootstrap values less than 70% within one species were clustered. Numbers in parentheses refer to the number of sequences include in each cluster. Species abbreviations: Adialytus ambiguus (ADamb), Aphidius avenae (APave), Aphidius colemani (APcol), Aphidius ervi (APerv), Aphidius matricariae (APmat), Aphidius microlophii (APmic), Aphidius rhopalosiphi (APrho), Aphidius uzbekistanicus (APuzb), Binodoxys angelicae (BIang), Diaeretiella rapae (DIrap), Ephedrus persicae (EPper) Ephedrus plagiator (EPpla), Lipolexis gracilis (LIgra), Lysiphlebus fabarum (LYfab), Lysiphlebus testaceipes (LYtes), Monoctonus crepidis (MOcre), Praon abjectum (PRabj), Praon gallicum (PRgal), Praon necans (PRnec), Praon volucre (PRvol), Toxares deltiger (TOdel), Trioxys auctus (TRaus) and Trioxys sp. A (TRspA). Last letter in the specimen code indicates whether sequencing was done in both directions (“B”) or just one direction (“S”—sense strand, “A”—antisense stand).