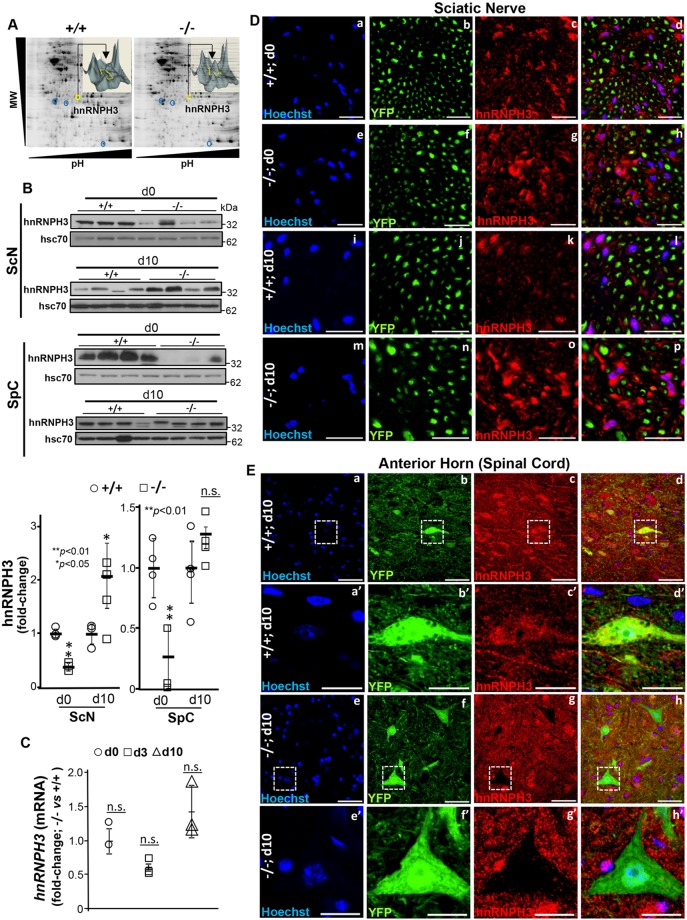
Fig. 8.
Post-transcriptional deregulation of hnRNPH3 in sciatic nerve and neural cell bodies by loss of Ranbp2. (A) Proteomic analyses by 2D-DIGE of homogenates of optic nerves of +/+ and −/− mice at d10. Insets are volumetric analyses of spot intensities marked by the yellow circle and arrow and that show an increase of a protein, which was identified by mass spectrometry as hnRNPH3, between +/+ and −/− mice. (B) Immunoblots (top panel) of equal amounts of homogenates and quantitative analyses (bottom panel) showing dysregulation of hnRNPH3 proteostasis in sciatic nerve (ScN) and spinal cord (SpC) at d0 and d10. *P<0.05, **P<0.01, Student's t-test, n=4 mice/genotype. Data are expressed as mean±s.d. Hsc70 is a loading control. For SpC d0, the same blots were used for P-Stat3, Stat3, hnRNPH3, Mmp28 and hsc70 in Figs 7-9. A statistical outlier was excluded in the dataset of hnRNPH3 in ScN at d0. Lanes shown are experimental replicates. (C) RT-qPCR showing that the temporal and transcriptional profile of hnRNPH3 mRNA in the sciatic nerve is not changed between +/+ and −/− mice. Student's t-test, n=4 mice/genotype. Data are expressed as mean±s.d. (D,E) Representative confocal images of cross-sections of sciatic nerve (D) and spinal cord (E) immunostained for hnRNPH3. (D) YFP+Hoechst−-axons lack hnRNPH3 and hnRNPH3 is localized in YFP− Hoechst+-Schwann cells, which have enhanced immunostaining of hnRNPH3 in −/− mice at d10. (E) The soma of YFP+ motoneurons in −/− anterior horns conspicuously lack hnRNPH3. Images of a′-d′ and e′-h′ are magnified views of the outlined regions in a-d and e-h, respectively. −/−, SLICK-H::Ranbp2flox/flox; +/+, SLICK-H::Ranbp2+/++/+; hsc70, heat shock protein 70; d0 and d10 are days 0 and 10 post-tamoxifen administration, respectively. Scale bars: 50 µm in Da-p,Ea-h; 20 µm in Ea′-h′; n.s., non-significant.