Fig. 3. Various protein structures aligned using the full protein’s heavy atoms, showing the rotation of the αC helix and of the glutamic acid residue (Glu46 in PDB 3G5D).
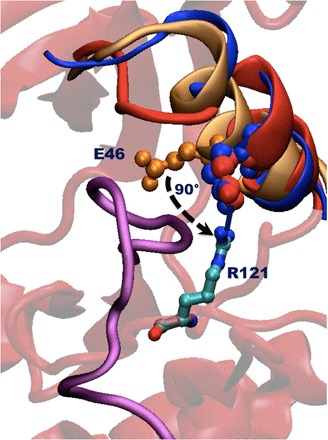
Orange represents equilibrated αC helix–in structure, red is the αC helix–out structure (or state 3 in Fig. 1) obtained from our runs, and blue is the αC helix–out structure directly from the PDB database (PDB 4YBK) as reported by Kwarcinski et al. (15). Note how the αC helices in the two out structures overlap fairly well and especially how the glutamic residue is displaced almost identically in a roughly orthogonal direction to what it had in the αC helix–in structure. The stabilizing interaction between residues Arg121 and Glu46 is also illustrated, along with the disordered activation loop in purple. The underlying base structure is a representative αC helix–out structure obtained from our runs. The reported structure (PDB 3G5D) in the protein database has missing residues after the end of the helix, and as such, we do not include them in the comparison here.
