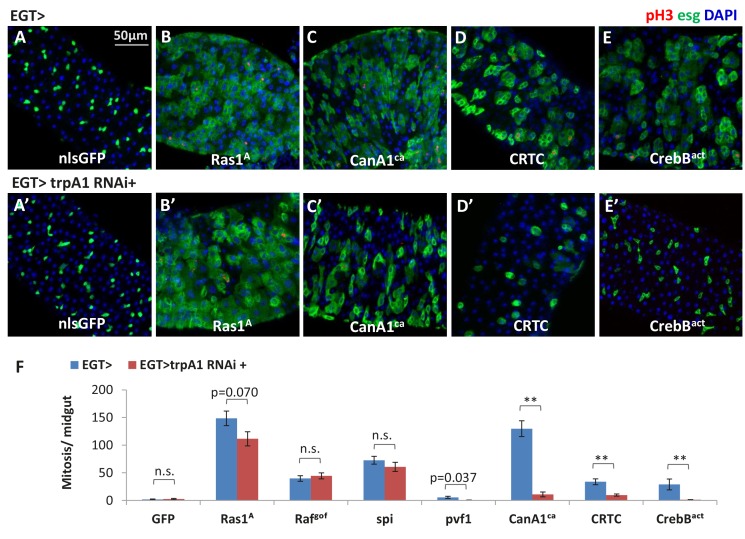
Figure 7. Ras/MAPK activity, not CanA/CRTC/CrebB, is sufficient for ISC proliferation in the absence of TRPA1.
(A–E) Midguts over-expressing calcium responsive signaling molecules such as active Ras1 (Ras1A), constitutively active Calcineurin A1 (CanA1ca), CREB-regulated transcription coactivator (CRTC), or active form of Creb (Crebact) in ISCs for 5d are stained for the mitosis marker pH3. The expansion of esg>GFP signal by CanA1ca is reduced by trpA1 RNAi, although not quite down to the wild-type level. This could be due to the different kinetics of two reagents, as CanA1ca may take effect sooner than trpA1 RNAi. (A’–E’) Midguts expressing trpA1 RNAi together with calcium-responsive signaling molecules in ISCs for 5d are stained for the mitosis marker pH3. (F) Mitosis quantification of midguts expressing GFP, Ras/ Raf, RTK ligands Spi/ Pvf1, CanA1ca, CRTC, Crebact, alone or together with trpA1 RNAi for genetic epistasis analysis. N > 5 midguts are analyzed for each genotype. Data are represented as mean ± SEM. Although it has been reported that pvf1 overexpression can increase ISC population (Bond and Foley, 2012), we could barely detect mitotic effect of Pvf1 in young adult flies.
DOI: http://dx.doi.org/10.7554/eLife.22441.031
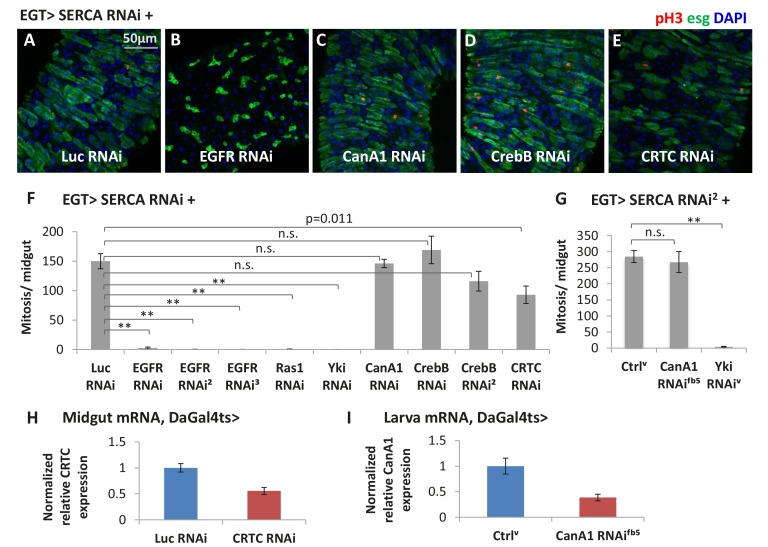
Figure 7—figure supplement 1. Ras/MAPK activity, but not CanA1/CrebB, is required for ISC proliferation induced by calcium influx.
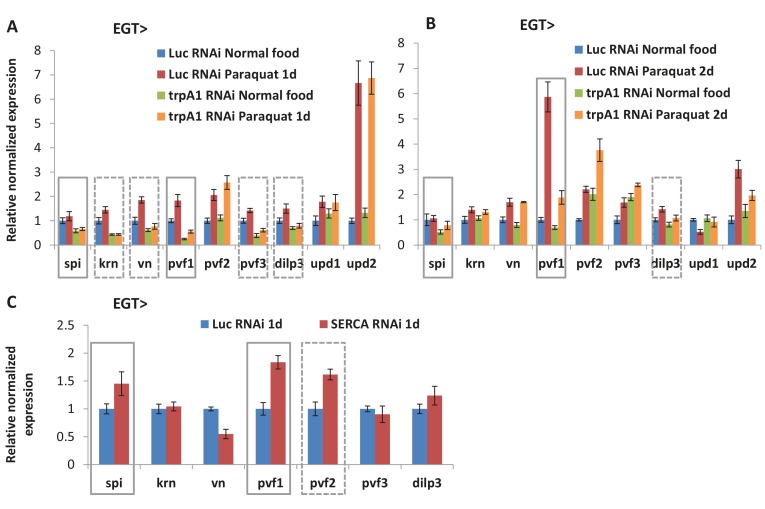
Figure 7—figure supplement 2. Ligands for receptor tyrosine kinases (RTKs) are affected by cytosolic Ca2+ signaling.
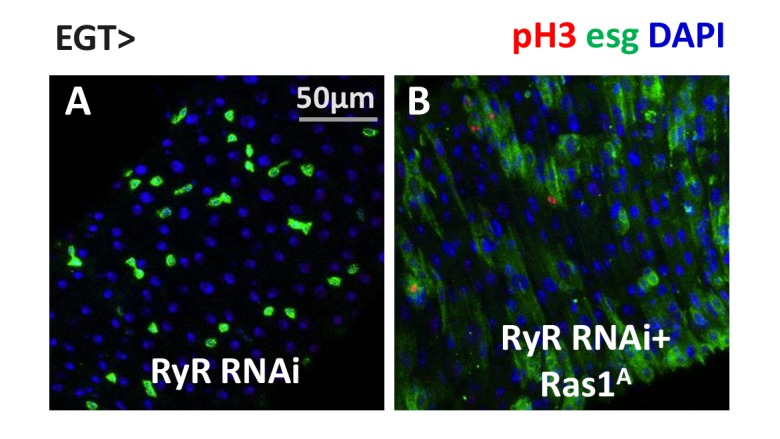
Figure 7—figure supplement 3. Ras/MAPK activity is sufficient for ISC proliferation in the absence of RyR.