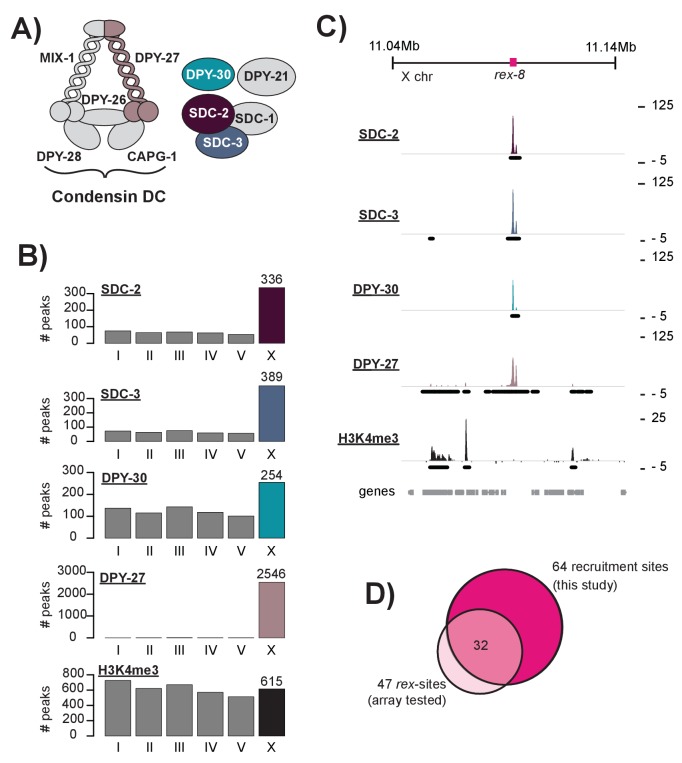
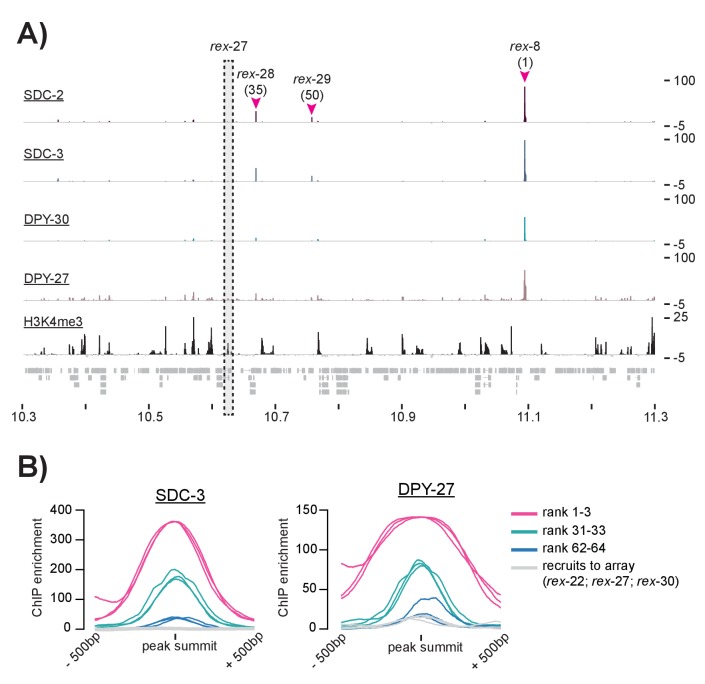
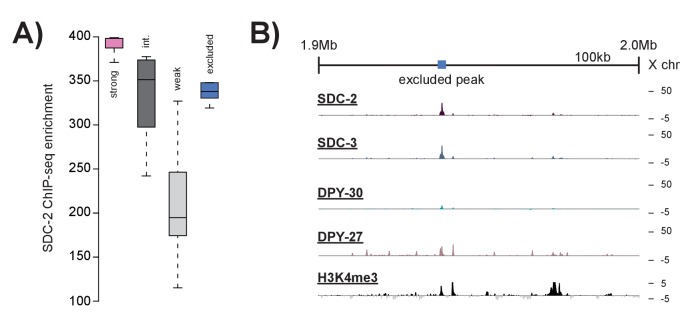
Figure 1. DCC recruitment sites are defined using high resolution ChIP-seq analysis.
(A) The C. elegans dosage compensation complex (DCC) is composed of a modified condensin complex (condensin DC) which is distinguished from condensin I by the SMC-4 variant, DPY-27. The non-condensin subunits SDC-2, SDC-3, and DPY-30 are required for DCC recruitment to the X chromosomes. (B) Peak distribution across each of the six chromosomes. (C) Representative ChIP-seq enrichment for an 80 kb window that includes recruitment element on the X, rex-8. Recruitment sites on the X chromosome were defined as a 400 bp window centered on the SDC-2 ChIP-seq summits that overlap with SDC-3, DPY-30, and DPY-27 peaks and that do not show significant H3K4me3 enrichment. (D) Recruitment sites identified in this study (n = 64) largely overlap with sequences previously shown to recruit DCC to multi-copy extrachromosomal array (n = 47) (Jans et al., 2009; Pferdehirt et al., 2011; McDonel et al., 2006).