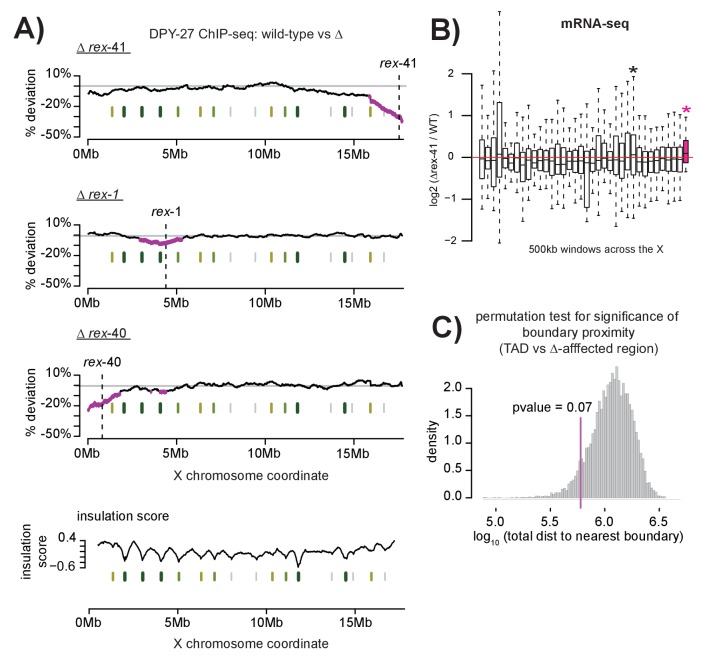
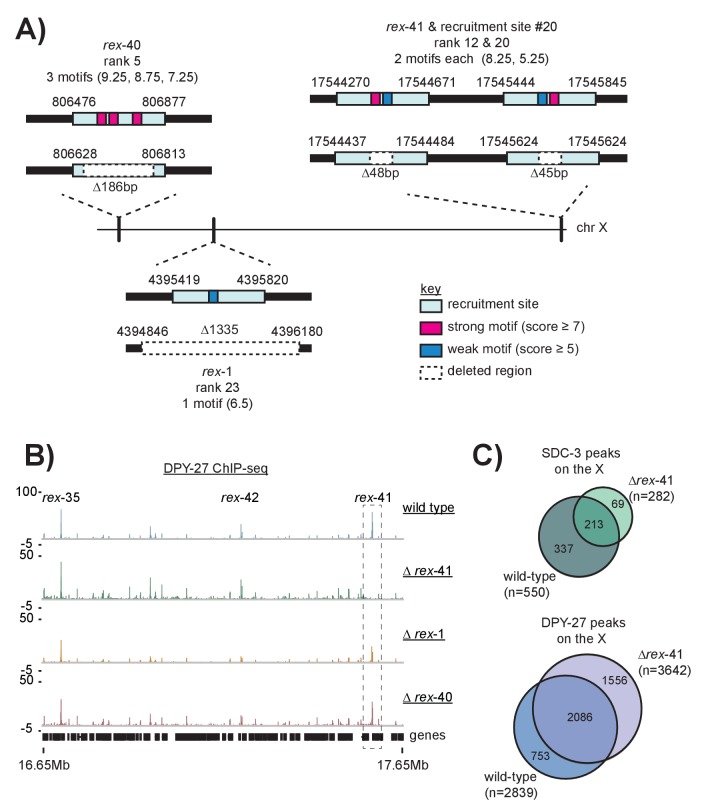
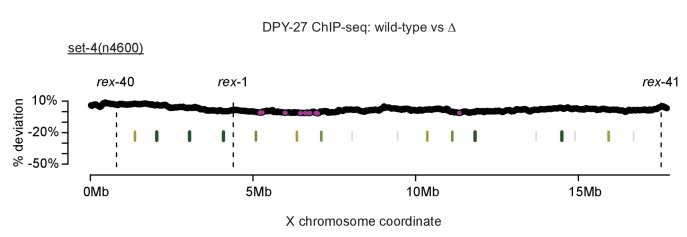
Figure 7. Multiple DCC recruitment sites establish DCC binding within defined X chromosomal domains.
(A) DPY-27 ChIP-seq data was used to calculate percent deviation from wild-type in three different recruitment site deletion strains. Briefly, the log2 ratio between wild-type and deletion strain was calculated for all DPY-27 ChIP-seq peaks and compared to a positive control locus (rex-8, the strongest recruitment site). A variable-step sliding window (window size of 2 Mb stepped across each DPY-27 peak) was used to calculate average deviation from wild-type binding. Regions shown in pink have significantly decreased DPY-27 enrichment compared to the rest of the chromosome, p-value<0.05, determined by one tailed students t-test, comparing log2 ratios in an individual window to log2 ratios across the whole X chromosome. In each experiment, the deleted recruitment site is indicated by a dashed line. Green lines (darker with stronger insulation activity) indicate TAD boundaries (as described in [Crane et al., 2015]). (B) mRNA-seq data comparing rex-41 deletion strain to wild-type is shown. Boxplot indicates the log2 ratio between deletion and wild-type for 500 kb windows tiled contiguously across the X chromosome. The rightmost window, shown in pink, contains a significant number of genes with increased transcription compared to wild-type (fisher test, p-value=0.0126). Asterisks mark windows with p-value<0.05). (C) Summed distance between borders of deletion-affected regions from all three recruitment site deletions in (A) and nearest TAD boundary (min: 54 kb, max: 230 kb) is closer than expected by chance alone (average: 313 kb). Shuffling the boundaries of deletion-affected regions (10000 permutations) produced a normal distribution wherein the observed proximity to TAD borders is expected to occur less than 7% of the time by chance. Shown is the distribution of the summed total distance between permuted deletion boundaries and nearest TAD boundary expressed as a log10 value.