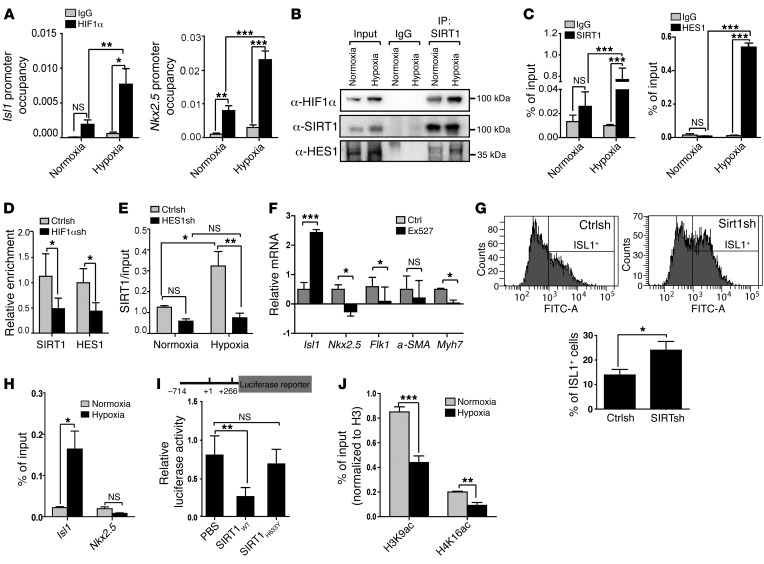
Figure 3. A SIRT1-HES1–containing complex represses Isl1 expression in a hypoxia-dependent manner.
(A) ChIP analysis of HIF1α at Isl1 (positions –468 to –285) (n = 4) and Nkx2.5 (positions –9040 to –8859) (n = 3) promoters after hypoxia treatment of day-6 EBs (1% O2, 16 hours). *P < 0.05; **P < 0.01; ***P < 0.001, ANOVA with Tukey’s post hoc correction. Enrichment of HIF1α was normalized to input DNA. (B) Co-IP assay of SIRT1 with HIF1α or HES1 under hypoxia. 5% input was used as loading control. 2 independent experiments were performed, generating similar results. (C) ChIP analyses of SIRT1 and HES1 binding to the Isl1 promoter (positions –468 to –285) under hypoxia (1% O2, 12 hours) and normoxia (21% O2) in E8.5 embryos. Enrichment of SIRT1 or HES1 was normalized to input DNA. ***P < 0.001, ANOVA with Tukey’s post hoc correction (n = 3). (D) ChIP analyses of SIRT1 (n = 8) and HES1 (n = 6) binding to the Isl1 promoter (positions –468 to –285) after lentivirus-mediated Hif1a knockdown in CoCl2–treated differentiating ESCs. Enrichment of SIRT1 or HES1 was normalized to input DNA. *P < 0.05, ANOVA with Tukey’s post hoc correction. (E) ChIP analysis of SIRT1 binding to the promoter of Isl1 (positions –468 to –285) in CPCs after Hes1 knockdown. *P < 0.05; **P < 0.01, ANOVA with Tukey’s post hoc correction (n = 3). (F) RT-qPCR analysis of Isl1, Nkx2.5, Flk1 (EB at E6), a-SMA, and Myh7 (EB at E8) expression in differentiating ES cells after treatment with the SIRT1 inhibitor (1 μM EX527, 24 hours; Isl1, Nkx2.5, a-SMA, Myh7: n = 3; Flk1: n = 5). The m34b4 gene was used as a reference for normalization. *P < 0.05; ***P < 0.001, t test. SIRT1 inhibitor treatment increases Isl1, but decreases Nkx2.5 and Flk1 expression. (G) FACS analyses of ISL1+ cells in EBs 6 days after Sirt1 knockdown. Left panels, histograms of ISL1+ cells. Right panel, quantification of ISL1+ cells. *P < 0.05, t test (n = 3). (H) ChIP analysis of SIRT1 binding to Isl1 (positions –468 to –285) and Nkx2.5 promoters (positions –9040 to –8859) in embryonic hearts after induction of hypoxia responses. *P < 0.05, t test (n = 3). (I) Luciferase reporter assays of the proximal Isl1 promoter with WT and H633Y mutant SIRT1. *P < 0.05; **P < 0.01, ANOVA with Dunnett’s post hoc correction (n = 3). (J) ChIP analysis of H3K9ac and H4K16ac at the Isl1 promoter in NKX2.5–EmGFP+ cells under hypoxia conditions. Enrichment of H3K9ac and histone H4K16ac was normalized to histone H3. **P < 0.01; ***P < 0.001, t test (n = 3).