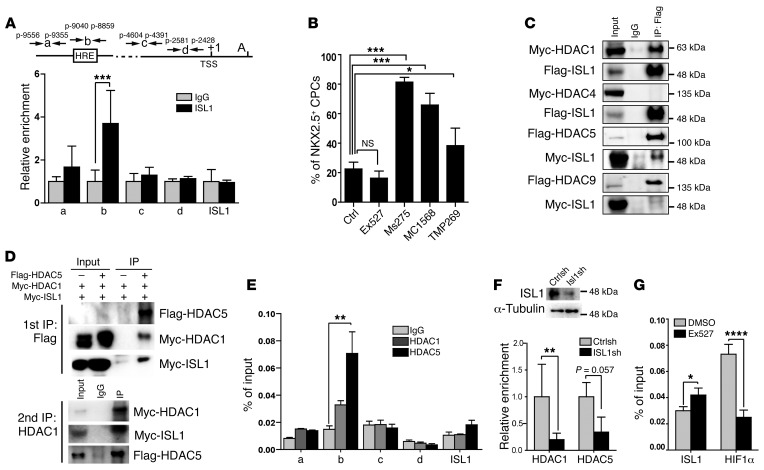
Figure 6. ISL1 represses Nkx2.5 transcription by recruitment of HDACs.
(A) ChIP analysis of ISL1 binding to the Nkx2.5 promoter. ***P < 0.001, t test (amplicons a–c, n = 4; amplicon d and ISL1, n = 3). Both the HRE motif and the putative ISL1-binding site are located at –9040 to –8859 (amplicon b). (B) Quantification of NKX2.5hi cells following in vitro differentiation of ISL1+ cells after treatment with HDAC inhibitors. *P < 0.05; ***P < 0.001, ANOVA with Dunnett’s post hoc correction (control, n = 6; Ex527, n = 5; Ms275 and MC1568, n = 3; TMP269, n = 5). (C) Co-IP analysis of the interaction of ISL1 with different HDACs in HEK293T cells after transfection. Two independent experiments were performed, generating similar results. (D) Sequential Co-IP analysis of the interaction of ISL1 with HDAC1 and HDAC5 in HEK293T cells. (E) ChIP analysis of HDAC1 and HDAC5 binding to the ISL1 site in Nkx2.5 promoter. **P < 0.01, ANOVA with Dunnett’s post hoc correction (amplicons a, c, d, ISL1, n = 3; amplicon b, n = 6). (F) ChIP analysis showing HDAC1 and HDAC5 binding to the Nkx2.5 promoter (positions –9040 to –8859) after lentiviral-mediated expression of ISL1 shRNA. **P < 0.01, t test (n = 3). (G) ChIP analysis showing ISL1 and HIF1α binding to the Nkx2.5 promoter (positions –9040 to –8859) after inhibition of SIRT1 enzymatic activity by Ex527 (1 μM). Note that enzymatic inhibition of SIRT1 enhances binding of ISL1, but reduces binding of HIF1α. *P < 0.05; ****P < 0.0001, t test (n = 3).