Figure 2.
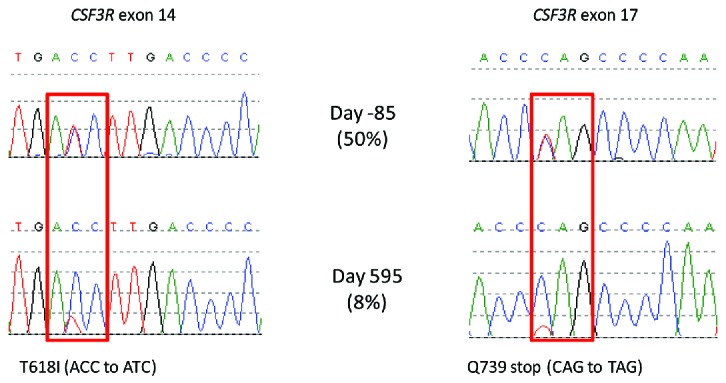
Different CSF3R mutations and their initial responses to treatment. CSF3R exons 14 and 17 were amplified from total peripheral blood leukocyte DNA and Sanger sequenced.5 To quantify the level of the T618I mutation, we designed a droplet digital PCR (ddPCR) assay. The ddPCR reaction mixture consisted of 10μl ddPCR supermix (Bio-Rad, Hemel Hempstead, UK), 250nm wild type probe ([HEX]ACCC[T]GA[T]GA[C]CT[T]GACC[BHQ1]), and mutation specific probe ([6FAM]ACCC[T]GA[T]GA[T]CT[T]GACC[BHQ1]), 900nm forward (CCACCAACAGTACAGTCC) and reverse (ACCAGGGGATTCAAAGTC) primers, and 7.2μl of fragmented DNA at 16.25ng/μl (bases in square brackets are locked nucleic acids). The entire reaction was loaded into a cartridge (Bio-Rad) together with 70μl droplet generation oil (Bio-Rad) and placed into the droplet generator (Bio-Rad). After processing, the droplets were transferred to a 96 well plate which was sealed with a heat sealer. PCR amplification was performed in a Thermal Cycler (Applied Biosystems 2720) using the following cycling conditions: 95°C for 10 mins, 40 cycles of 94°C for 30s and 60.8C for 1 min, 1 cycle of 98°C for 10 mins and ending at 4°C. After amplification, the plate was loaded onto the droplet reader (Bio-Rad) and analysed immediately with Quantasoft analysis software (Bio-Rad).
