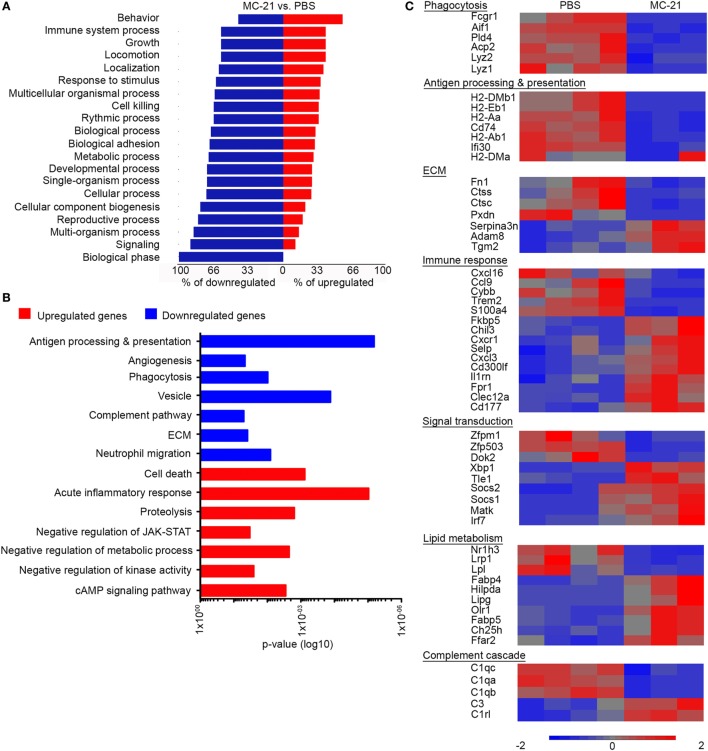
Figure 5.
Altered neutrophil-mediated innate immune functions and cell survival in the absence of Ly6Chi monocytes. (A) Forest plot analysis of 449 differentially expressed genes (p < 0.05; t-test, with at least twofold change) presenting for each biological process the percentage of downregulated genes (in blue) and upregulated genes (in red). (B) Graphic representation of the significance (p-value) for the enrichment of selected GO categories out of DAVID analyses performed for the downregulated (in blue) and upregulated genes (in red) comparing between “MC-21” versus “phosphate-buffered saline (PBS)” neutrophils. Analysis was performed on the differentially expressed genes (p < 0.05; t-test, ≥2-fold change). (C) RNA-seq heat map analysis displaying the differential expression (z-scored) of genes between “MC-21” and “PBS” neutrophils, which are associated with phagocytosis, antigen processing and presentation, extracellular matrix (ECM) remodeling, immune response, signal transduction, lipid metabolism, and complement cascade. Color legend is presented at the bottom of the figure. Analysis was performed on the differentially expressed genes (p < 0.05; t-test, ≥2-fold change).