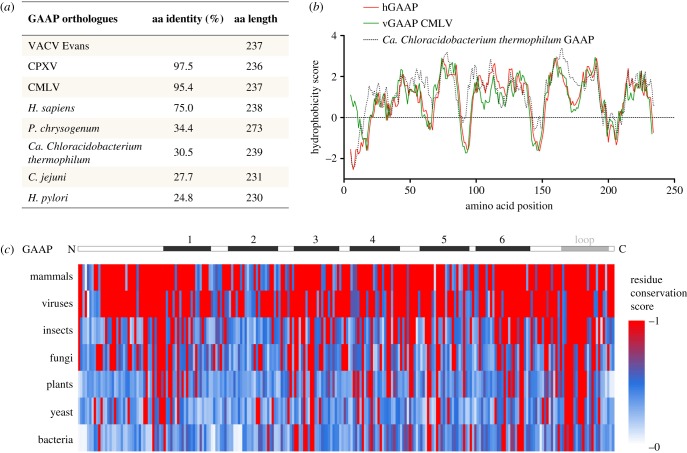
Figure 1.
Conservation within the GAAPs. The extent of hydrophobicity and sequence conservation among GAAP orthologues. (a) The aa identities calculated by the BLASTP server and differences in aa length are indicated. (b) The hydrophobicity profile for hGAAP (eukaryote) was aligned with that of viral (vGAAP from CMLV) and prokaryotic (Ca. Chloracidobacterium thermophilum) GAAP representatives. Complete aa sequences were used for all. (c) aa sequence alignment of hGAAP against GAAP orthologues from two to three representative members from each taxon. The level of conservation for each residue was scored according to Scorecons and represented in a colour gradient, with red and white indicating identity and no similarity, respectively. Sequences analysed include Homo sapiens, Bos taurus and Gallus gallus (vertebrates); VACV Evans, CMLV and CPXV (viruses); Cerapachys biroi and Tribolium castaneum (insects); Penicillium chrysogenum and Tuber melanosporum (fungi); Arabidopsis thaliana, Genlisea aurea and Zea mays (plants); Schizosaccharomyces pombe and Saccharomyces cerevisiae (yeast); and Campylobacter jejuni, Helicobacter pylori and Candidatus Chloracidobacterium thermophilum (bacteria). Black and grey boxes indicate the location of TMDs 1–6 and the hydrophobic region/loop 7 of GAAPs, respectively. Adapted from Carrara et al. [4].