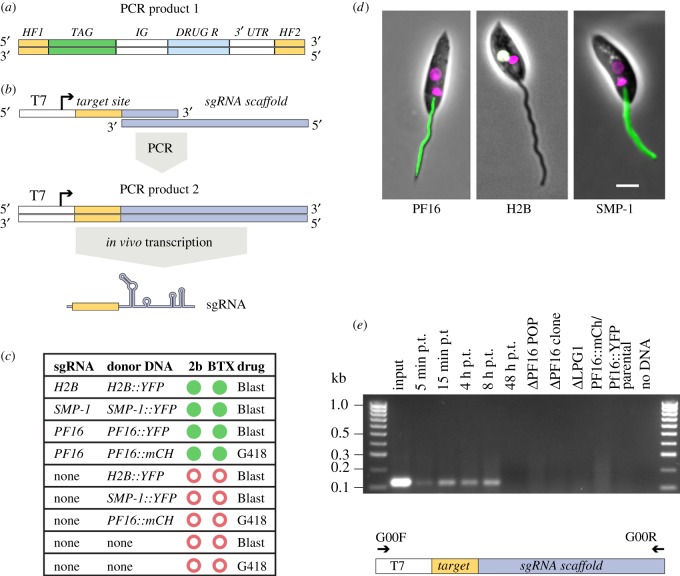
Figure 3.
Co-transfection of two PCR amplicons allowed precise insertion of marker genes. (a) PCR-amplified donor DNA containing 30 nt HF specific to the target locus, a fluorescent protein tag and a drug-selectable marker gene. (b) Strategy for sgRNA delivery: the sgRNA template is produced by PCR using an oligo encoding the T7 promoter, 20 nt defining the target-site and a sequence complementary to the 3′-end of the second oligo, comprising the sgRNA scaffold [28]. The resulting PCR product is transfected into cells for T7 RNAP-driven transcription of the sgRNA. (c) Summary of outcome of transfections with different combinations of sgRNA templates and donor DNAs and electroporation protocols. Green filled circles denote drug-resistant cells showing the expected fluorescent signal; red open circles indicate failure to produce any drug-resistant transfectants. (d) Micrographs showing correct flagellar localization of PF16::YFP, nuclear localization of H2B::YFP (the white colour indicates co-localization of YFP and Hoechst) and flagellar membrane localization of SMP-1::YFP in cells that were co-transfected with the donor DNA and corresponding sgRNA template. Phase contrast image merged with mCh or YFP fluorescence channels and Hoechst-stained DNA (magenta). Scale bar 5 µm. (e) PCR-detection of the sgRNA template. Top, agarose gel showing the results of a diagnostic PCR to test for the presence of the sgRNA template. Template DNAs were as follows. Input: 1 µl of PF16 sgRNA PCR used for transfection; 5 min-48 h post transfection (p.t.): genomic DNA from cells at different time points post transfection with PF16 sgRNA; ΔPF16, ΔLPG1, PF16::mCh / PF16::YFP: genomic DNA from drug-resistant cell lines reported in this study; parental: genomic DNA from the parental cell line L. mex Cas9 T7. Bottom, diagram showing the sgRNA template and the primers used for PCR detection.