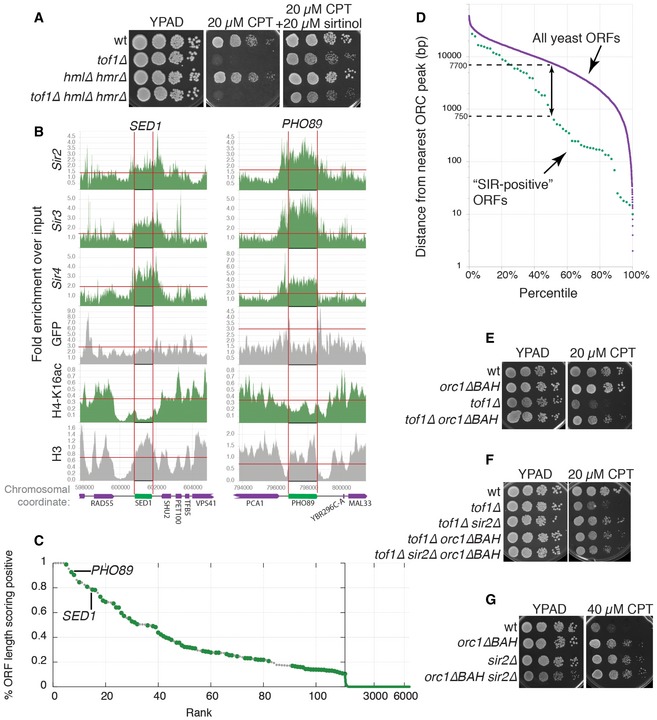
Deletion of HML and HMR does not suppress camptothecin hypersensitivity of tof1∆ strains.
Analysis of ChIP‐seq data for the proteins indicated on the y‐axes. Enrichments are plotted as a function of the genomic coordinate; in green is the protein/modification tested; in grey are controls. The ORFs indicated at the top of each graph are identified by two vertical red lines. The horizontal red lines indicate the thresholds used in this work to determine enrichment (Sir2/Sir3/Sir4/GFP/H3) or loss (H4‐K16) of ChIP signal.
Identification of regions bound by the SIR complex: for each ORF in the genome, a “SIR score” was calculated as the fraction of the ORF for which both increased Sir2, Sir3, Sir4, and decreased H4‐K16ac were observed. ORFs were sorted based of their “SIR score”. Subtelomeric ORFs and ORFs proximal to HML and HMR are shown with small grey dots, while remaining ORFs are shown with large green dots.
SIR‐positive ORFs are on average located closer to sites of ORC binding than ORFs in general. All yeast ORFs are shown in purple as a function of their distance from the nearest site of ORC binding. Non‐telomeric and non‐HM SIR‐positive ORFs (SIR score > 0.2) are shown in green.
Deletion of the BAH domain of ORC1 partially rescues camptothecin hypersensitivity of tof1∆ cells.
In a tof1∆orc1∆BAH background, deletion of SIR2 does not further increase camptothecin resistance.
SIR2 deletion and orc1∆BAH mutation are epistatic with regard to camptothecin resistance.