Figure 1. ZBTB48 binds telomeres via its ZnF11 domain in vitro and in vivo .
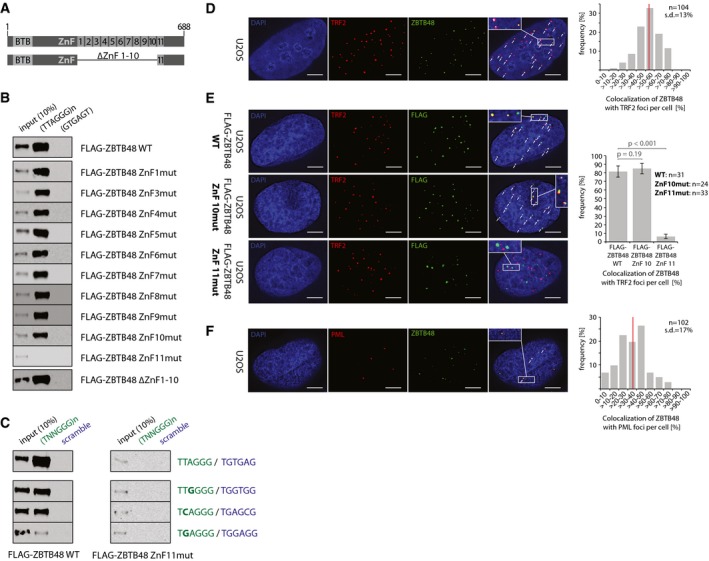
- Domain structure of wild‐type (WT) ZBTB48 containing a N‐terminal BTB domain and 11 zinc fingers (ZnF) at the C‐terminus of the 688‐amino‐acid protein. Note that ZnF2 is degenerate. Below is a schematic of the deletion construct lacking ZnF1‐10 used in (B).
- Sequence‐specific DNA pull‐downs with either telomeric (TTAGGG) or a control sequence (GTGAGT) for FLAG‐ZBTB48 WT, point mutants for the ten functional zinc fingers and a domain deletion construct for ZnF1‐10.
- Sequence‐specific DNA pull‐downs for FLAG‐ZBTB48 WT and ZnF11 point mutant for telomeric and subtelomeric variant repeat sequences (green) and their respective scrambled controls (blue).
- Co‐localization analysis of endogenous ZBTB48 and TRF2 in U2OS cells by immunofluorescence (IF) staining. A representative image illustrating the co‐localization between ZBTB48 (green) and TRF2 (red) as a marker for telomeres is shown with DAPI (blue) used as a nuclear counterstain. The quantification of frequency of co‐localization events (right) was done after 3D reconstruction of the acquired z‐stacks (n = 104 cells). The average value is indicated by a red bar.
- IF stainings for exogenous FLAG‐ZBTB48 WT and point mutants for ZnF10 and ZnF11 in U2OS cells. The same analysis as in (D) was performed and average co‐localization frequencies are shown (n = 24–33 cells). Error bars indicate standard deviations, and P‐values are based on Student's t‐test.
- Co‐localization analysis between ZBTB48 (green) and PML (red) as a marker for PML bodies analogous to (D) in U2OS cells (n = 102 cells).
