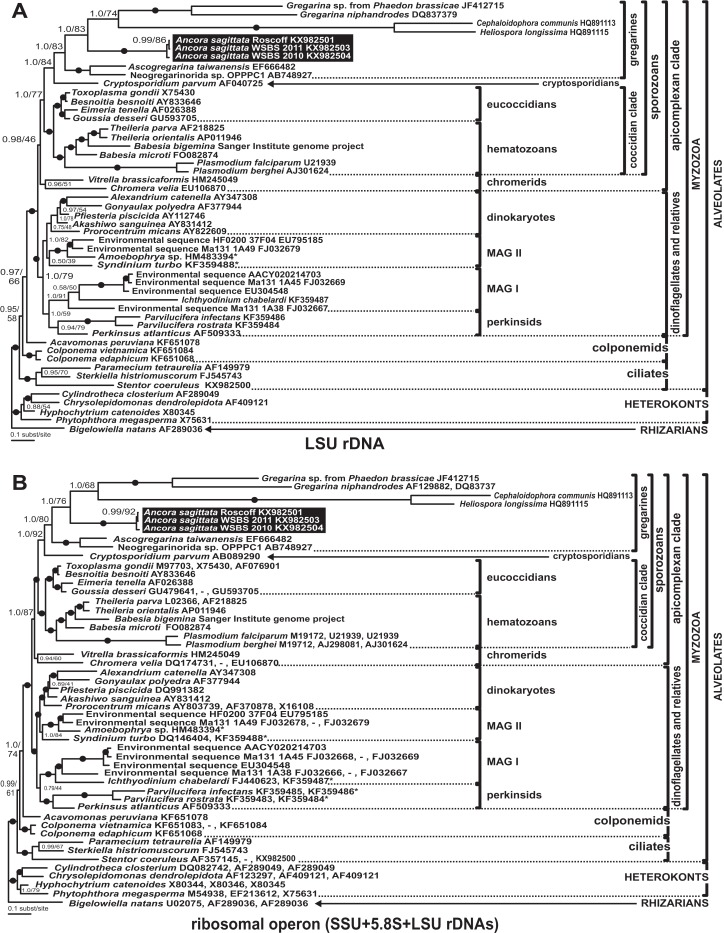
Figure 9. Bayesian inference trees of the alveolates obtained by using the GTR+Г+I model and 50 sequences.
(A) LSU rDNA dataset (2,911 sites); (B) Ribosomal operon dataset (4,636 sites). Numbers at the nodes indicate Bayesian posterior probabilities/ML bootstrap percentages. Black dots on the branches indicate Bayesian posterior probabilities and bootstrap percentages of at least 95% and 95%, respectively. The newly obtained sequences of Ancora sagittata are highlighted by black rectangles. Accession numbers in (B) are arranged in following order: SSU rDNA, 5.8S (if available), LSU rDNA. The sequences of Babesia bigemina were obtained from the Sanger Institute genome project (https://www.sanger.ac.uk/Projects/B_bigemina/). Asterisks mark partial LSU rDNA sequences of small size (300–700 bp).