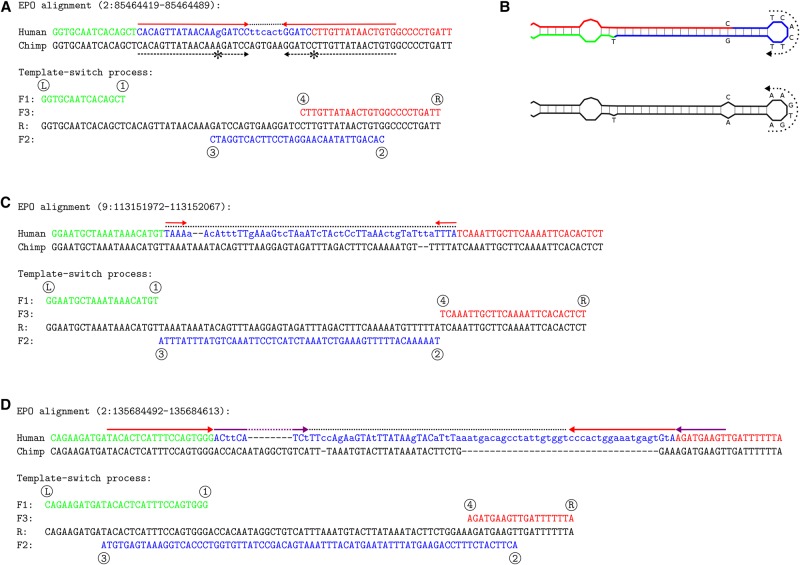
Figure 2.
Example events detected in human. (A) A near-perfect inverted repeat in chimp (dashed black arrows, the one mismatch indicated with asterisks) has been converted into a perfect inverted repeat (red arrows) in human (top). The cluster of six additional dissimilarities (dotted line) in fact represents perfect inversion of the 6-bp spacer sequence and makes the template switch (bottom) a likely explanation (Original data: http://grch37.ensembl.org/Homo_sapiens/Location/Compara_Alignments?align=548;r=2:85464419-85464489). (B) Predicted DNA secondary structure before (chimp; bottom) and after (human; top) the template switch event. The dotted arrows indicate the reverse-complemented spacer region, which the four-point model explains with a single event. (C,D) Additional complex mutation patterns (mismatches in lower case) that can be explained by a single template switch event. (C) Event “1-3-2-4” only converts the spacer sequence (http://grch37.ensembl.org/Homo_sapiens/Location/Compara_Alignments?align=548;r=9:113151972-113152067). (D) Event “3-1-4-2” converts the spacer sequence and creates two inverted repeats (red and magenta arrows) (http://grch37.ensembl.org/Homo_sapiens/Location/Compara_Alignments?align=548;r=2:135684492-135684613).