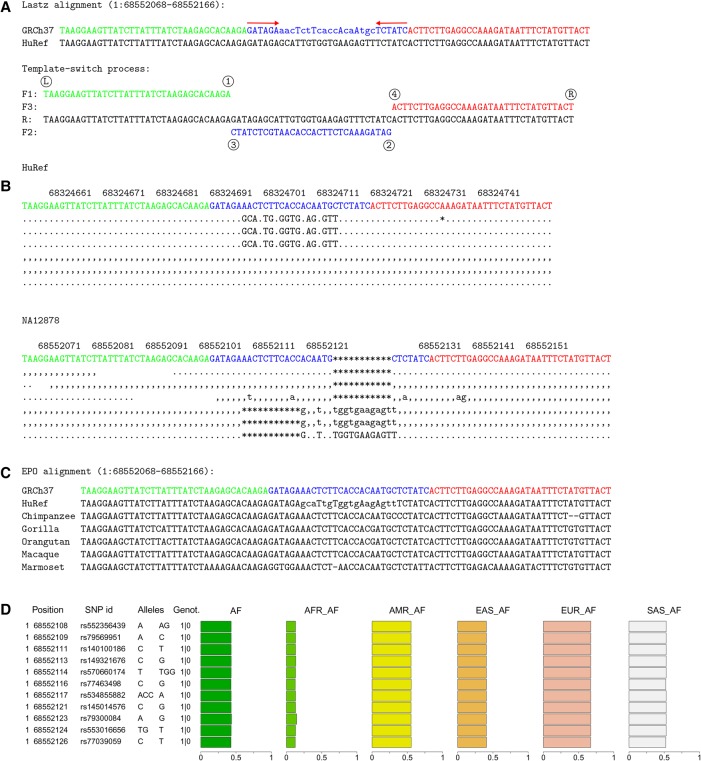
Figure 3.
A template switch mutation event with variable allele frequencies in human populations. (A) Four-point model explanation of a complex mutation between the human reference GRCh37 and HuRef. Notation is as in Figure 1. (B) A subset of the original sequencing reads from HuRef (top) and the 1KG individual NA12878 (bottom). Dots and commas indicate the read matching to the reference on the forward and reverse strand, uppercase and lowercase characters denote the corresponding mismatches, and asterisks mark the alignment gaps. These reads reveal heterozygosity at the locus. (C) The EPO alignment for primates reveals that GRCh37 is the ancestral form. As all other primates resemble the reference allele, the most parsimonious explanation is that the mutation (HuRef) happened in the human lineage since its divergence from the human–chimp ancestor. (D) 1KG variation data explain this event as a cluster of seven single-nucleotide polymorphisms and four indels. The phased genotypes for NA12878 (1|0) indicate that the variant alleles are linked and all in the same haplotype. The single origin of the whole cluster is further supported by the uniform derived allele frequencies across the sites within all 1KG data (AF) and within each superpopulation (AFR, AMR, EAS, EUR, SAS).