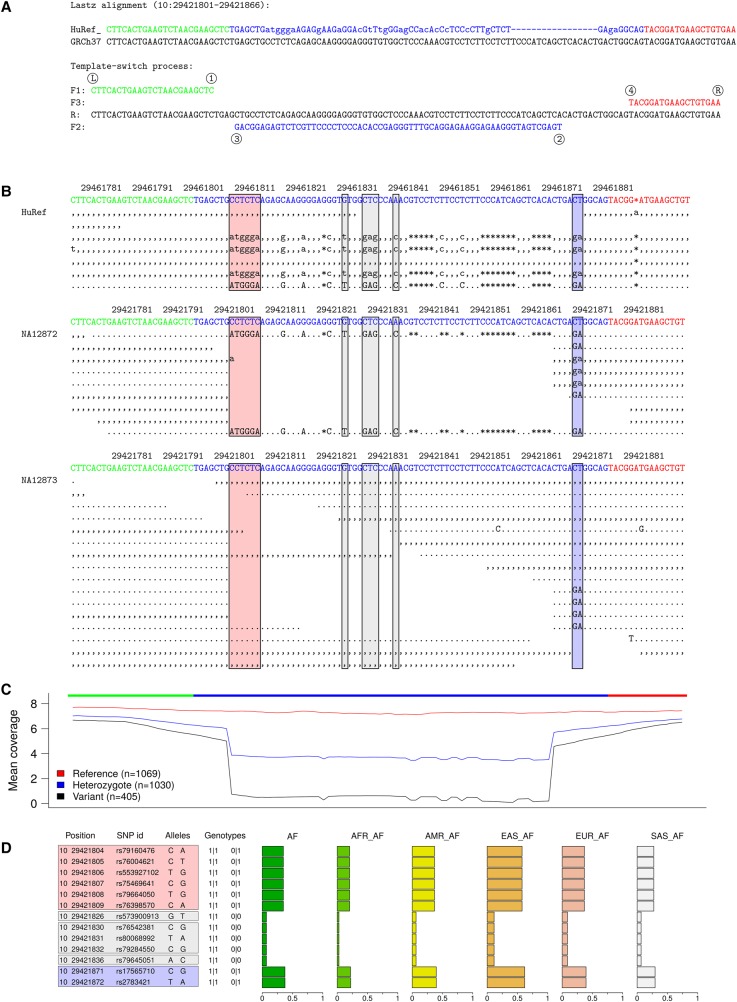
Figure 4.
Complex mutation partially called in 1KG data. (A) A mutation pattern explained by a template switch event. (B) Sequence reads from HuRef and 1KG individuals NA12872 and NA12873. Well-aligned capillary reads reveal HuRef as a heterozygote. In the 1KG individuals, the short NGS reads for the variant allele mostly fail to map: based on two indicative sites (blue box), NA12872 is a homozygote and NA12873 is a heterozygote. (C) Mean sequencing depth of all 2504 1KG (phase 3) low-coverage individuals, grouped according to their genotype for the two indicative sites. (D) 1KG variation data, with genotypes for NA12872 (left) and NA12873 (right). Adjacent variants created by a template switch event are transmitted together and are expected to have identical genotypes, giving uniform allele frequencies in population data. This is not the case here, because fewer than half of the differences (colored boxes, corresponding to those in B) are called, and of those, the terminal ones are called with a higher certainty, the mutation pattern is incomplete, and the variant alleles at adjacent positions appear to occur at different frequencies.